Note
Go to the end to download the full example code.
Create a Gaussian process¶
import openturns as ot
import openturns.viewer as viewer
from matplotlib import pylab as plt
ot.Log.Show(ot.Log.NONE)
Create a Gaussian process from a covariance model¶
In this paragraph we build a Gaussian process from its covariance model.
We first define a covariance model :
defaultDimension = 1
# Amplitude values
amplitude = [1.0] * defaultDimension
# Scale values
scale = [1.0] * defaultDimension
# Covariance model
myModel = ot.AbsoluteExponential(scale, amplitude)
We define a mesh,
tmin = 0.0
step = 0.1
n = 11
myTimeGrid = ot.RegularGrid(tmin, step, n)
and create the process :
process = ot.GaussianProcess(myModel, myTimeGrid)
print(process)
GaussianProcess(trend=[x0]->[0.0], covariance=AbsoluteExponential(scale=[1], amplitude=[1]))
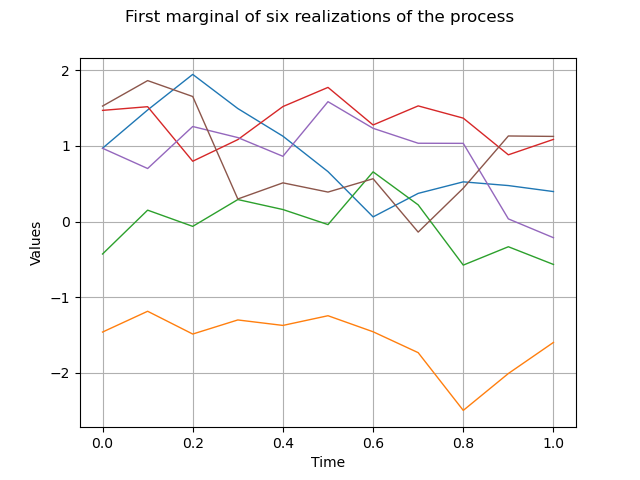
We draw the first marginal of a sample of size 6 :
sample = process.getSample(6)
graph = sample.drawMarginal(0)
graph.setTitle("First marginal of six realizations of the process")
view = viewer.View(graph)
Create a Gaussian process from spectral density¶
In this paragraph we build a Gaussian process from its spectral density.
We first define a spectral model :
amplitude = [1.0, 2.0]
scale = [4.0, 5.0]
spatialCorrelation = ot.CorrelationMatrix(2)
spatialCorrelation[0, 1] = 0.8
mySpectralModel = ot.CauchyModel(scale, amplitude, spatialCorrelation)
As usual we define a mesh,
myTimeGrid = ot.RegularGrid(0.0, 0.1, 20)
and create the process thereafter
process = ot.SpectralGaussianProcess(mySpectralModel, myTimeGrid)
print(process)
SpectralGaussianProcess=SpectralGaussianProcess dimension=2 spectralModel=class=CauchyModel amplitude=[1,2] scale=[4,5] spatial correlation=
[[ 1 0.8 ]
[ 0.8 1 ]] maximal frequency=5 n frequency=10
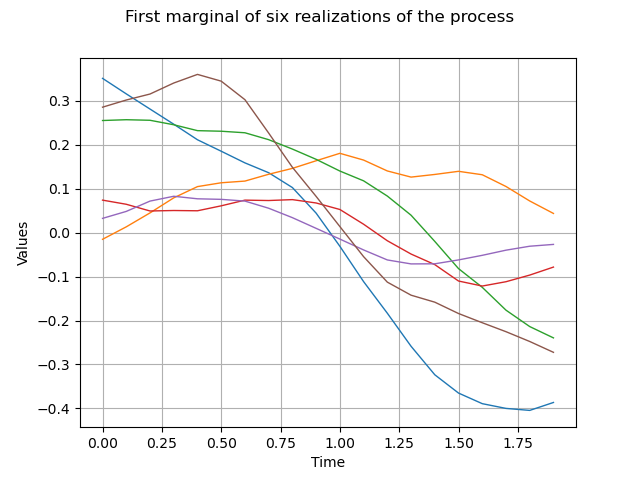
Eventually we draw the first marginal of a sample of size 6 :
sample = process.getSample(6)
graph = sample.drawMarginal(0)
graph.setTitle("First marginal of six realizations of the process")
view = viewer.View(graph)
Display figures
plt.show()
 OpenTURNS
OpenTURNS