Note
Click here to download the full example code
Estimate a non stationary covariance function¶
The objective of this use case is to estimate from several
fields generated by the process
. We suppose that the process
is not stationary.
In the following example, we illustrate the estimation of the non stationary covariance model
defined by:
The domain is discretized on a mesh
which is a time grid with 64 points.
We build a normal process
with zero mean and
as covariance function.
We discretize the covariance model
using
for each
.
We get a
fields from the process
from wich we estimate the covariance model
.
We use the object NonStationaryCovarianceModelFactory which creates a UserDefinedCovarianceModel.
from __future__ import print_function
import math as m
import openturns as ot
import openturns.viewer as viewer
from matplotlib import pylab as plt
ot.Log.Show(ot.Log.NONE)
Create the time grid
t0 = -4.0
tmax = 4.0
N = 64
dt = (tmax - t0) / N
tgrid = ot.RegularGrid(t0, dt, N)
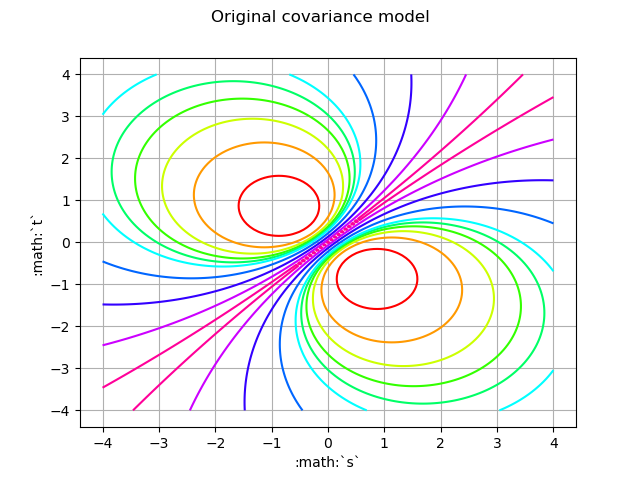
Create the covariance function at (s,t)
def C(s, t):
return m.exp(-4.0 * abs(s - t) / (1 + (s * s + t * t)))
Draw…
def f(X):
s, t = X
return [C(s, t)]
func = ot.PythonFunction(2, 1, f)
func.setDescription([':math:`s`', ':math:`t`', ':math:`cov`'])
graph = func.draw([t0] * 2, [tmax] * 2)
graph.setTitle('Original covariance model')
graph.setLegendPosition('')
view = viewer.View(graph)
Create data from a non stationary normal process Omega * [0,T]–> R
# Create the collection of HermitianMatrix
covariance = ot.CovarianceMatrix(N)
for k in range(N):
s = tgrid.getValue(k)
for l in range(k + 1):
t = tgrid.getValue(l)
covariance[k, l] = C(s, t)
covmodel = ot.UserDefinedCovarianceModel(tgrid, covariance)
Create the normal process with that covariance model based on the mesh tgrid
process = ot.GaussianProcess(covmodel, tgrid)
# Get a sample of fields from the process
N = 1000
sample = process.getSample(N)
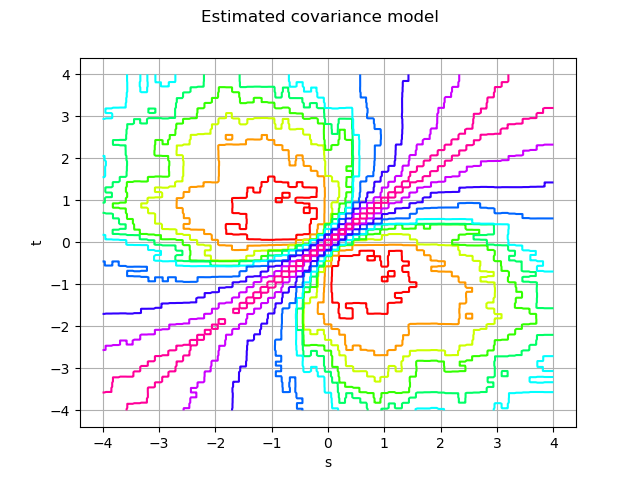
The covariance model factory
factory = ot.NonStationaryCovarianceModelFactory()
# Estimation on a sample
estimatedModel = factory.build(sample)
graph = estimatedModel.draw(0, 0, t0, tmax, 256, False)
graph.setTitle('Estimated covariance model')
graph.setLegendPosition('')
view = viewer.View(graph)
plt.show()
Total running time of the script: ( 0 minutes 0.266 seconds)
 OpenTURNS
OpenTURNS