Note
Click here to download the full example code
Bayesian calibration of the flooding model¶
Abstract¶
The goal of this example is to present the statistical hypotheses of the bayesian calibration of the flooding model.
Parameters to calibrate¶
The vector of parameters to calibrate is:
The variables to calibrate are and are set to the following values:
Observations¶
In this section, we describe the statistical model associated with the observations.
The errors of the water heights are associated with a gaussian distribution with a zero mean and a standard variation equal to:
Therefore, the observed water heights are:
for where
and we make the hypothesis that the observation errors are independent. We consider a sample size equal to:
The observations are the couples , i.e. each observation is a couple made of the flowrate and the corresponding river height.
Analysis¶
In this model, the variables and
are not identifiables, since only the difference
matters. Hence, calibrating this model requires some regularization.
Generate the observations¶
import pylab as pl
from openturns.viewer import View
from openturns.usecases import flood_model as flood_model
import openturns.viewer as viewer
import numpy as np
import openturns as ot
ot.Log.Show(ot.Log.NONE)
A basic implementation of the probabilistic model is available in the usecases module :
fm = flood_model.FloodModel()
We define the model which has 4 inputs and one output H.
The nonlinear least squares does not take into account for bounds in the parameters. Therefore, we ensure that the output is computed whatever the inputs. The model fails into two situations:
if
,
if
.
In these cases, we return an infinite number.
def functionFlooding(X):
L = 5.0e3
B = 300.0
Q, K_s, Z_v, Z_m = X
alpha = (Z_m - Z_v)/L
if alpha < 0.0 or K_s <= 0.0:
H = np.inf
else:
H = (Q/(K_s*B*np.sqrt(alpha)))**(3.0/5.0)
return [H]
g = ot.PythonFunction(4, 1, functionFlooding)
g = ot.MemoizeFunction(g)
g.setOutputDescription(["H (m)"])
We load the input distribution for .
Q = fm.Q
Set the parameters to be calibrated.
K_s = ot.Dirac(30.0)
Z_v = ot.Dirac(50.0)
Z_m = ot.Dirac(55.0)
K_s.setDescription(["Ks (m^(1/3)/s)"])
Z_v.setDescription(["Zv (m)"])
Z_m.setDescription(["Zm (m)"])
We create the joint input distribution.
inputRandomVector = ot.ComposedDistribution([Q, K_s, Z_v, Z_m])
Create a Monte-Carlo sample of the output H.
nbobs = 20
inputSample = inputRandomVector.getSample(nbobs)
outputH = g(inputSample)
Generate the observation noise and add it to the output of the model.
sigmaObservationNoiseH = 0.1 # (m)
noiseH = ot.Normal(0., sigmaObservationNoiseH)
ot.RandomGenerator.SetSeed(0)
sampleNoiseH = noiseH.getSample(nbobs)
Hobs = outputH + sampleNoiseH
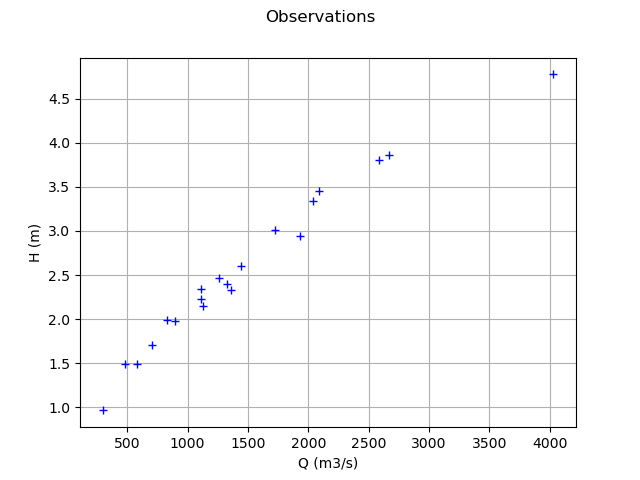
Plot the Y observations versus the X observations.
Qobs = inputSample[:, 0]
graph = ot.Graph("Observations", "Q (m3/s)", "H (m)", True)
cloud = ot.Cloud(Qobs, Hobs)
graph.add(cloud)
view = viewer.View(graph)
Setting the calibration parameters¶
Define the parametric model that associates each observation
and values of the parameters
to the parameters of the distribution of the corresponding observation: here
def fullModelPy(X):
Q, K_s, Z_v, Z_m = X
H = g(X)[0]
sigmaH = 0.5 # (m^2) The standard deviation of the observation error.
return [H, sigmaH]
fullModel = ot.PythonFunction(4, 2, fullModelPy)
model = ot.ParametricFunction(fullModel, [0], Qobs[0])
model
Define the value of the reference values of the parameter. In the bayesian framework, this is called the mean of the prior gaussian distribution. In the data assimilation framework, this is called the background.
KsInitial = 20.
ZvInitial = 49.
ZmInitial = 51.
parameterPriorMean = [KsInitial, ZvInitial, ZmInitial]
paramDim = len(parameterPriorMean)
Define the covariance matrix of the parameters to calibrate.
sigmaKs = 5.
sigmaZv = 1.
sigmaZm = 1.
parameterPriorCovariance = ot.CovarianceMatrix(paramDim)
parameterPriorCovariance[0, 0] = sigmaKs**2
parameterPriorCovariance[1, 1] = sigmaZv**2
parameterPriorCovariance[2, 2] = sigmaZm**2
parameterPriorCovariance
Define the the prior distribution of the parameter
prior = ot.Normal(parameterPriorMean, parameterPriorCovariance)
prior.setDescription(['Ks', 'Zv', 'Zm'])
prior
Define the distribution of observations conditional on model predictions.
Note that its parameter dimension is the one of , so the model must be adjusted accordingly. In other words, the input argument of the setParameter method of the conditional distribution must be equal to the dimension of the output of the model. Hence, we do not have to set the actual parameters: only the type of distribution is used.
conditional = ot.Normal()
conditional
Proposal distribution: uniform.
proposal = [ot.Uniform(-5., 5.), ot.Uniform(-1., 1.), ot.Uniform(-1., 1.)]
proposal
Out:
[class=Uniform name=Uniform dimension=1 a=-5 b=5, class=Uniform name=Uniform dimension=1 a=-1 b=1, class=Uniform name=Uniform dimension=1 a=-1 b=1]
Test the MCMC sampler¶
The MCMC sampler essentially computes the log-likelihood of the parameters.
mymcmc = ot.MCMC(prior, conditional, model, Qobs, Hobs, parameterPriorMean)
mymcmc.computeLogLikelihood(parameterPriorMean)
Out:
-146.3520105560049
Test the Metropolis-Hastings sampler¶
Creation of the Random Walk Metropolis-Hastings (RWMH) sampler.
initialState = parameterPriorMean
RWMHsampler = ot.RandomWalkMetropolisHastings(
prior, conditional, model, Qobs, Hobs, initialState, proposal)
Tuning of the RWMH algorithm.
Strategy of calibration for the random walk (trivial example: default).
strategy = ot.CalibrationStrategyCollection(paramDim)
RWMHsampler.setCalibrationStrategyPerComponent(strategy)
Other parameters.
RWMHsampler.setVerbose(True)
RWMHsampler.setThinning(1)
RWMHsampler.setBurnIn(200)
Generate a sample from the posterior distribution of the parameters theta.
sampleSize = 1000
sample = RWMHsampler.getSample(sampleSize)
Look at the acceptance rate (basic checking of the efficiency of the tuning; value close to 0.2 usually recommended).
RWMHsampler.getAcceptanceRate()
Build the distribution of the posterior by kernel smoothing.
kernel = ot.KernelSmoothing()
posterior = kernel.build(sample)
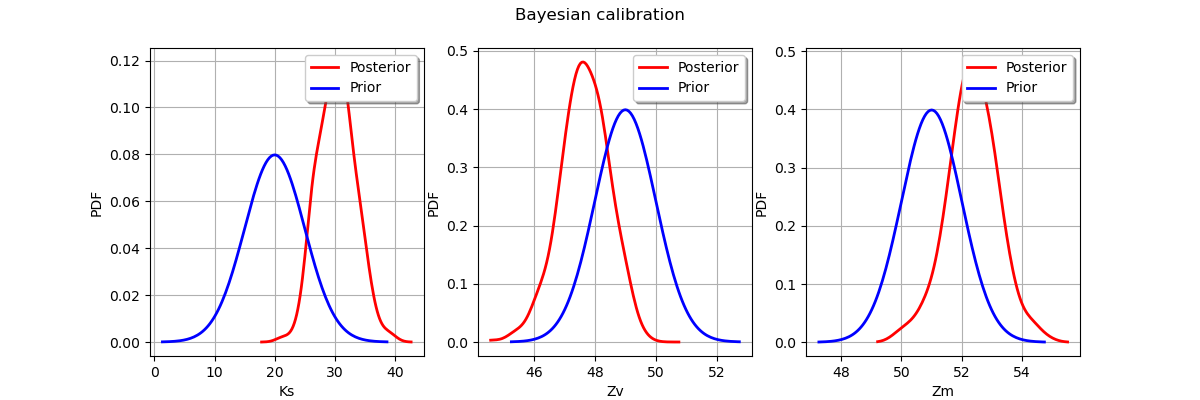
Display prior vs posterior for each parameter.
fig = pl.figure(figsize=(12, 4))
for parameter_index in range(paramDim):
graph = posterior.getMarginal(parameter_index).drawPDF()
priorGraph = prior.getMarginal(parameter_index).drawPDF()
priorGraph.setColors(['blue'])
graph.add(priorGraph)
graph.setLegends(['Posterior', 'Prior'])
ax = fig.add_subplot(1, paramDim, parameter_index+1)
_ = ot.viewer.View(graph, figure=fig, axes=[ax])
_ = fig.suptitle("Bayesian calibration")
Total running time of the script: ( 0 minutes 1.920 seconds)
 OpenTURNS
OpenTURNS