Note
Go to the end to download the full example code
Calibration of the logistic model¶
We present a calibration study of the logistic growth model defined here. In this example, we calibrate the parameters of a model which predicts the dynamics of the size of a population. This shows how you can calibrate a model which predicts a time dependent output. You need to view the output time series as a vector.
Analysis of the data¶
from openturns.usecases import logistic_model
import openturns as ot
import numpy as np
import openturns.viewer as viewer
from matplotlib import pylab as plt
ot.Log.Show(ot.Log.NONE)
We load the logistic model from the usecases module :
lm = logistic_model.LogisticModel()
The data is based on 22 dates from 1790 to 2000. The observation points are stored in the data field :
observedSample = lm.data
nbobs = observedSample.getSize()
nbobs
22
timeObservations = observedSample[:, 0]
timeObservations[0:5]
populationObservations = observedSample[:, 1]
populationObservations[0:5]
graph = ot.Graph("", "Time (years)", "Population (Millions)", True, "topleft")
cloud = ot.Cloud(timeObservations, populationObservations)
cloud.setLegend("Observations")
graph.add(cloud)
view = viewer.View(graph)
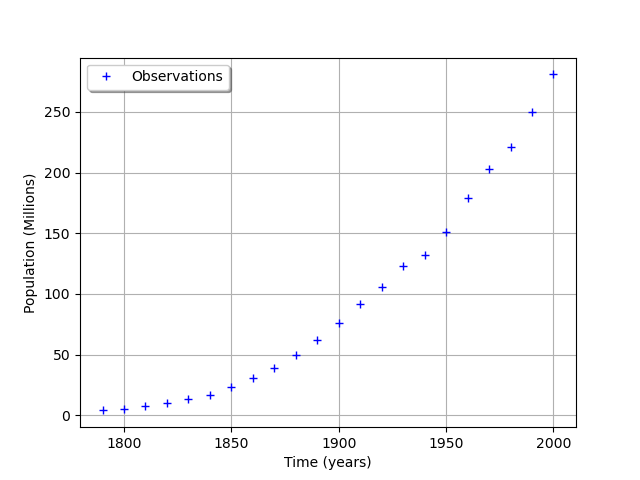
We consider the times and populations as dimension 22 vectors. The logisticModel function takes a dimension 24 vector as input and returns a dimension 22 vector. The first 22 components of the input vector contains the dates and the remaining 2 components are and
.
nbdates = 22
def logisticModel(X):
t = [X[i] for i in range(nbdates)]
a = X[22]
c = X[23]
t0 = 1790.0
y0 = 3.9e6
b = np.exp(c)
y = [0.0] * nbdates
for i in range(nbdates):
y[i] = a * y0 / (b * y0 + (a - b * y0) * np.exp(-a * (t[i] - t0)))
z = [yi / 1.0e6 for yi in y] # Convert into millions
return z
logisticModelPy = ot.PythonFunction(24, nbdates, logisticModel)
The reference values of the parameters.
Because is so small with respect to
, we use the logarithm.
np.log(1.5587e-10)
-22.581998789427587
a = 0.03134
c = -22.58
thetaPrior = [a, c]
logisticParametric = ot.ParametricFunction(logisticModelPy, [22, 23], thetaPrior)
Check that we can evaluate the parametric function.
populationPredicted = logisticParametric(timeObservations.asPoint())
populationPredicted
graph = ot.Graph("", "Time (years)", "Population (Millions)", True, "topleft")
# Observations
cloud = ot.Cloud(timeObservations, populationObservations)
cloud.setLegend("Observations")
cloud.setColor("blue")
graph.add(cloud)
# Predictions
cloud = ot.Cloud(timeObservations.asPoint(), populationPredicted)
cloud.setLegend("Predictions")
cloud.setColor("green")
graph.add(cloud)
view = viewer.View(graph)
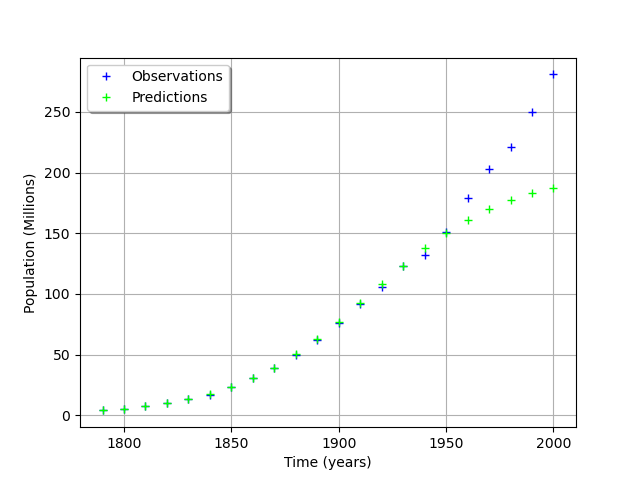
We see that the fit is not good: the observations continue to grow after 1950, while the growth of the prediction seem to fade.
Calibration with linear least squares¶
timeObservationsVector = ot.Sample([[timeObservations[i, 0] for i in range(nbobs)]])
timeObservationsVector[0:10]
populationObservationsVector = ot.Sample(
[[populationObservations[i, 0] for i in range(nbobs)]]
)
populationObservationsVector[0:10]
The reference values of the parameters.
a = 0.03134
c = -22.58
thetaPrior = [a, c]
logisticParametric = ot.ParametricFunction(logisticModelPy, [22, 23], thetaPrior)
Check that we can evaluate the parametric function.
populationPredicted = logisticParametric(timeObservationsVector)
populationPredicted[0:10]
Calibration¶
algo = ot.LinearLeastSquaresCalibration(
logisticParametric, timeObservationsVector, populationObservationsVector, thetaPrior
)
algo.run()
calibrationResult = algo.getResult()
thetaMAP = calibrationResult.getParameterMAP()
thetaMAP
thetaPosterior = calibrationResult.getParameterPosterior()
thetaPosterior.computeBilateralConfidenceIntervalWithMarginalProbability(0.95)[0]
transpose samples to interpret several observations instead of several input/outputs as it is a field model
if calibrationResult.getInputObservations().getSize() == 1:
calibrationResult.setInputObservations(
[timeObservations[i] for i in range(nbdates)]
)
calibrationResult.setOutputObservations(
[populationObservations[i] for i in range(nbdates)]
)
outputAtPrior = [
[calibrationResult.getOutputAtPriorMean()[0, i]] for i in range(nbdates)
]
outputAtPosterior = [
[calibrationResult.getOutputAtPosteriorMean()[0, i]] for i in range(nbdates)
]
calibrationResult.setOutputAtPriorAndPosteriorMean(outputAtPrior, outputAtPosterior)
graph = calibrationResult.drawObservationsVsInputs()
view = viewer.View(graph)
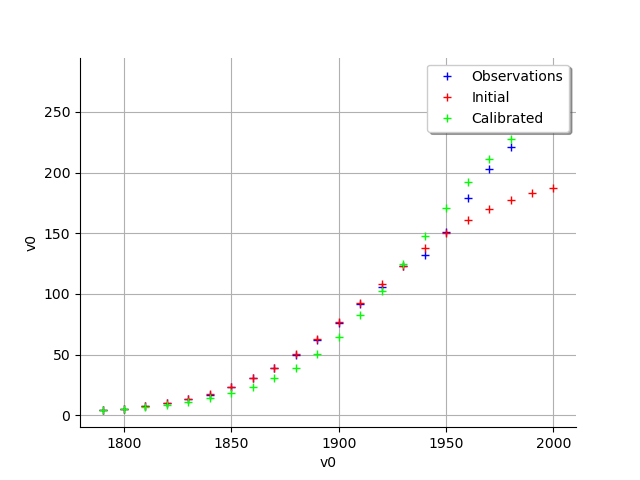
graph = calibrationResult.drawObservationsVsInputs()
view = viewer.View(graph)
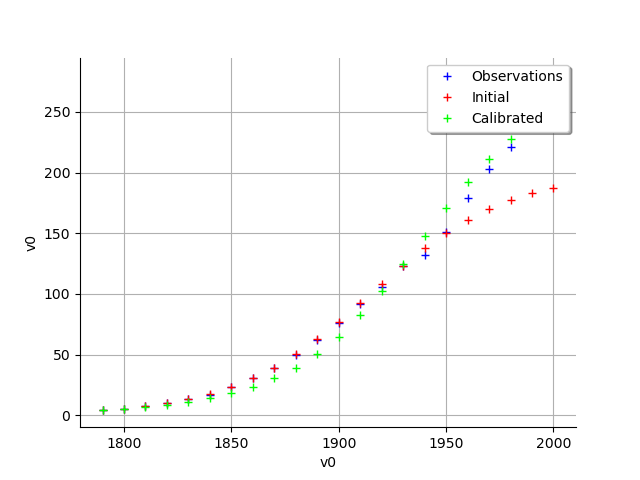
graph = calibrationResult.drawObservationsVsPredictions()
view = viewer.View(graph)
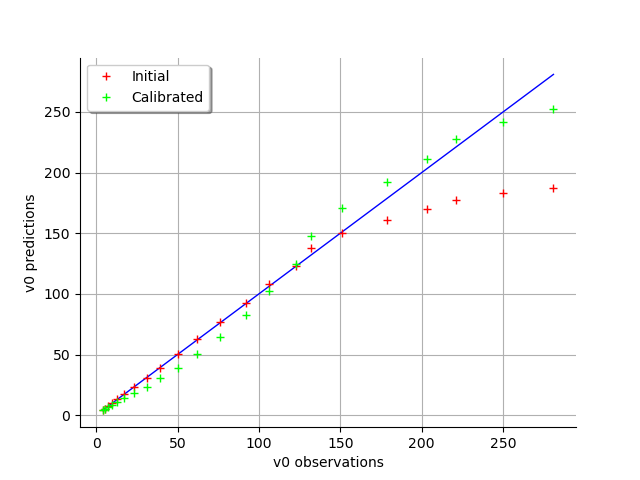
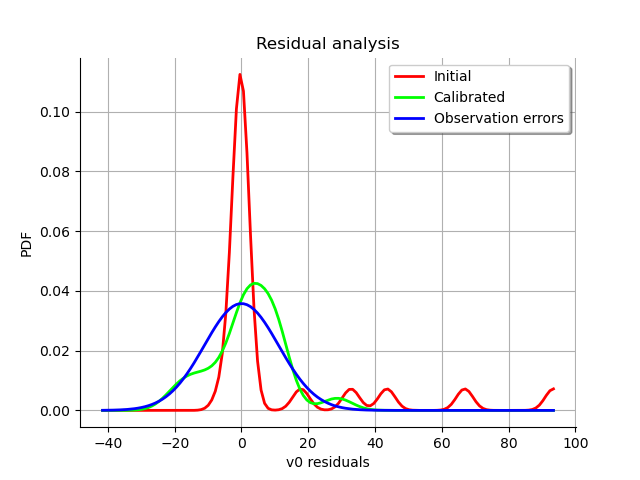
graph = calibrationResult.drawResiduals()
view = viewer.View(graph)
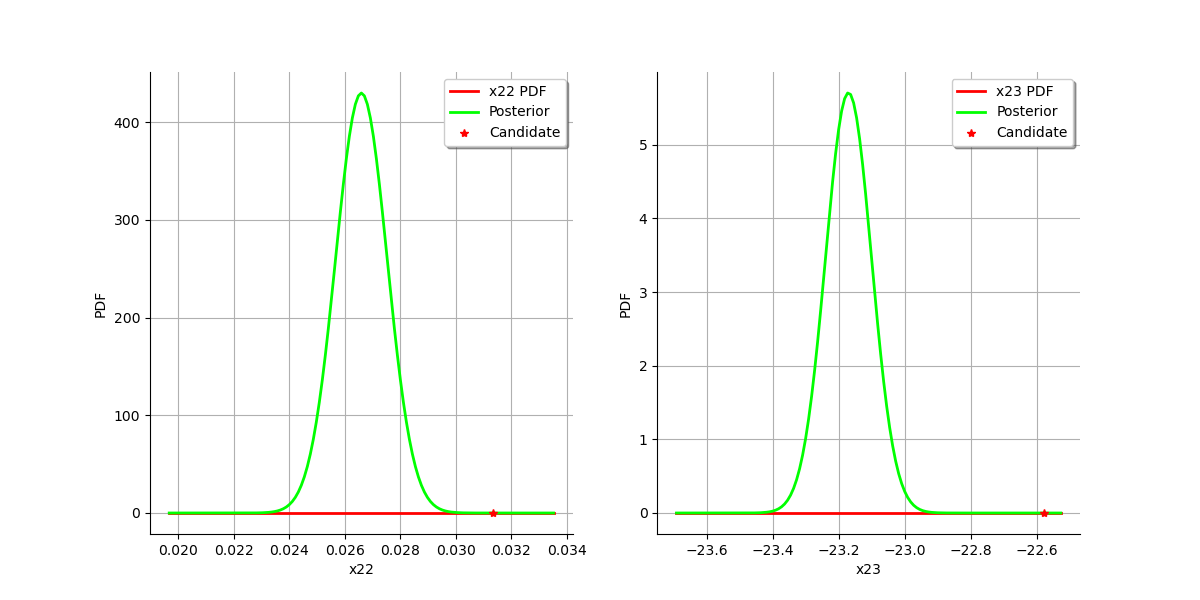
graph = calibrationResult.drawParameterDistributions()
view = viewer.View(graph)
plt.show()
Total running time of the script: ( 0 minutes 0.851 seconds)
 OpenTURNS
OpenTURNS