Note
Go to the end to download the full example code
Create a stationary covariance model¶
This use case illustrates how the user can define his own stationary covariance model thanks to the object UserDefinedStationaryCovarianceModel defined from:
a mesh
of dimension
defined by the vertices
and the associated simplices,
a collection of covariance matrices stored in the object CovarianceMatrixCollection noted
where
for
Then we build a stationary covariance function which is a piecewise constant function on defined by:
where is such that
is the vertex of
the nearest to
import openturns as ot
import openturns.viewer as viewer
from matplotlib import pylab as plt
ot.Log.Show(ot.Log.NONE)
We detail the example described in the documentation Create the time grid
t0 = 0.0
dt = 0.5
N = int((20.0 - t0) / dt)
mesh = ot.RegularGrid(t0, dt, N)
# Create the covariance function
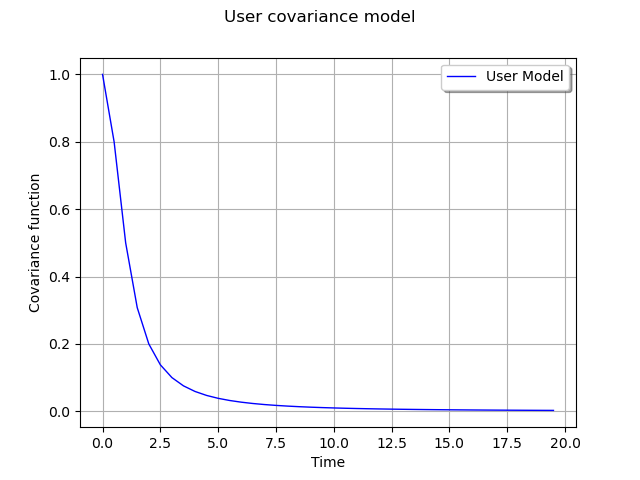
def gamma(tau):
return 1.0 / (1.0 + tau * tau)
# Create the collection of HermitianMatrix
coll = ot.SquareMatrixCollection()
for k in range(N):
t = mesh.getValue(k)
matrix = ot.SquareMatrix([[gamma(t)]])
coll.add(matrix)
Create the covariance model
covmodel = ot.UserDefinedStationaryCovarianceModel(mesh, coll)
# One vertex of the mesh
tau = 1.5
# Get the covariance function computed at the vertex tau
covmodel(tau)
Graph of the spectral function
x = ot.Sample(N, 2)
for k in range(N):
t = mesh.getValue(k)
x[k, 0] = t
value = covmodel(t)
x[k, 1] = value[0, 0]
# Create the curve of the spectral function
curve = ot.Curve(x, "User Model")
# Create the graph
myGraph = ot.Graph("User covariance model", "Time", "Covariance function", True)
myGraph.add(curve)
myGraph.setLegendPosition("topright")
view = viewer.View(myGraph)
plt.show()
Total running time of the script: ( 0 minutes 0.071 seconds)
 OpenTURNS
OpenTURNS