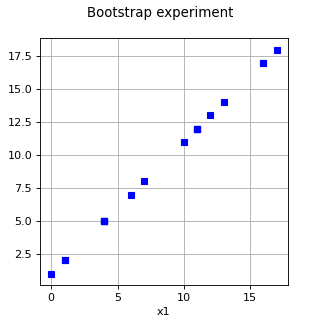
BootstrapExperiment¶
(Source code, png, hires.png, pdf)
- class BootstrapExperiment(*args)¶
Bootstrap experiment.
- Available constructors:
BootstrapExperiment(sample)
- Parameters:
- sample2-d sequence of float
Points to defined a
UserDefineddistribution.
See also
Notes
BootstrapExperiment is a random weighted design of experiments. Calling the
BootstrapExperimentconstructor is equivalent to calling theWeightedExperimentconstructor as follows:WeightedExperiment(UserDefined(sample), sample.getSize())Examples
>>> import openturns as ot >>> ot.RandomGenerator.SetSeed(0) >>> sample = [[i,i+1] for i in range(5)] >>> experiment = ot.BootstrapExperiment(sample) >>> print(experiment.generate()) [ v0 v1 ] 0 : [ 4 5 ] 1 : [ 1 2 ] 2 : [ 1 2 ] 3 : [ 1 2 ] 4 : [ 2 3 ] >>> print(experiment.getDistribution()) UserDefined({x = [0,1], p = 0.2}, {x = [1,2], p = 0.2}, {x = [2,3], p = 0.2}, {x = [3,4], p = 0.2}, {x = [4,5], p = 0.2})
Methods
GenerateSelection(size, length)Generate a list of indices of points with replacement.
generate()Generate points according to the type of the experiment.
Generate points and their associated weight according to the type of the experiment.
Accessor to the object's name.
Accessor to the distribution.
getId()Accessor to the object's id.
getName()Accessor to the object's name.
Accessor to the object's shadowed id.
getSize()Accessor to the size of the generated sample.
Accessor to the object's visibility state.
hasName()Test if the object is named.
Ask whether the experiment has uniform weights.
Test if the object has a distinguishable name.
setDistribution(distribution)Accessor to the distribution.
setName(name)Accessor to the object's name.
setShadowedId(id)Accessor to the object's shadowed id.
setSize(size)Accessor to the size of the generated sample.
setVisibility(visible)Accessor to the object's visibility state.
- __init__(*args)¶
- static GenerateSelection(size, length)¶
Generate a list of indices of points with replacement.
- Parameters:
- sizepositive int
Number of indices to choose.
- npositive int
Upper bound of the interval in which the indices are chosen.
- Returns:
- selection
Indices Sequence of size size of indices
such that
.
- selection
- generate()¶
Generate points according to the type of the experiment.
- Returns:
- sample
Sample Points
which constitute the design of experiments with
. The sampling method is defined by the nature of the weighted experiment.
- sample
Examples
>>> import openturns as ot >>> ot.RandomGenerator.SetSeed(0) >>> myExperiment = ot.MonteCarloExperiment(ot.Normal(2), 5) >>> sample = myExperiment.generate() >>> print(sample) [ X0 X1 ] 0 : [ 0.608202 -1.26617 ] 1 : [ -0.438266 1.20548 ] 2 : [ -2.18139 0.350042 ] 3 : [ -0.355007 1.43725 ] 4 : [ 0.810668 0.793156 ]
- generateWithWeights()¶
Generate points and their associated weight according to the type of the experiment.
- Returns:
Examples
>>> import openturns as ot >>> ot.RandomGenerator.SetSeed(0) >>> myExperiment = ot.MonteCarloExperiment(ot.Normal(2), 5) >>> sample, weights = myExperiment.generateWithWeights() >>> print(sample) [ X0 X1 ] 0 : [ 0.608202 -1.26617 ] 1 : [ -0.438266 1.20548 ] 2 : [ -2.18139 0.350042 ] 3 : [ -0.355007 1.43725 ] 4 : [ 0.810668 0.793156 ] >>> print(weights) [0.2,0.2,0.2,0.2,0.2]
- getClassName()¶
Accessor to the object’s name.
- Returns:
- class_namestr
The object class name (object.__class__.__name__).
- getDistribution()¶
Accessor to the distribution.
- Returns:
- distribution
Distribution Distribution used to generate the set of input data.
- distribution
- getId()¶
Accessor to the object’s id.
- Returns:
- idint
Internal unique identifier.
- getName()¶
Accessor to the object’s name.
- Returns:
- namestr
The name of the object.
- getShadowedId()¶
Accessor to the object’s shadowed id.
- Returns:
- idint
Internal unique identifier.
- getSize()¶
Accessor to the size of the generated sample.
- Returns:
- sizepositive int
Number
of points constituting the design of experiments.
- getVisibility()¶
Accessor to the object’s visibility state.
- Returns:
- visiblebool
Visibility flag.
- hasName()¶
Test if the object is named.
- Returns:
- hasNamebool
True if the name is not empty.
- hasUniformWeights()¶
Ask whether the experiment has uniform weights.
- Returns:
- hasUniformWeightsbool
Whether the experiment has uniform weights.
- hasVisibleName()¶
Test if the object has a distinguishable name.
- Returns:
- hasVisibleNamebool
True if the name is not empty and not the default one.
- setDistribution(distribution)¶
Accessor to the distribution.
- Parameters:
- distribution
Distribution Distribution used to generate the set of input data.
- distribution
- setName(name)¶
Accessor to the object’s name.
- Parameters:
- namestr
The name of the object.
- setShadowedId(id)¶
Accessor to the object’s shadowed id.
- Parameters:
- idint
Internal unique identifier.
- setSize(size)¶
Accessor to the size of the generated sample.
- Parameters:
- sizepositive int
Number
of points constituting the design of experiments.
- setVisibility(visible)¶
Accessor to the object’s visibility state.
- Parameters:
- visiblebool
Visibility flag.
 OpenTURNS
OpenTURNS