Note
Go to the end to download the full example code
Create a custom covariance model¶
This example illustrates how the user can define his own covariance model.
import openturns as ot
import openturns.viewer as viewer
from matplotlib import pylab as plt
import math as m
ot.Log.Show(ot.Log.NONE)
Create the time grid
N = 32
a = 4.0
mesh = ot.IntervalMesher([N]).build(ot.Interval(-a, a))
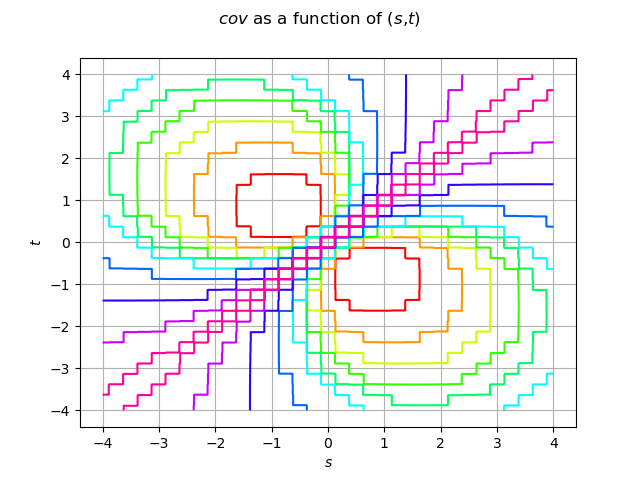
Create the covariance function at (s,t)
def C(s, t):
return m.exp(-4.0 * abs(s - t) / (1 + (s * s + t * t)))
Create the large covariance matrix
covariance = ot.CovarianceMatrix(mesh.getVerticesNumber())
for k in range(mesh.getVerticesNumber()):
t = mesh.getVertices()[k]
for ll in range(k + 1):
s = mesh.getVertices()[ll]
covariance[k, ll] = C(s[0], t[0])
Create the covariance model
covmodel = ot.UserDefinedCovarianceModel(mesh, covariance)
Draw the covariance model
def f(x):
return [covmodel([x[0]], [x[1]])[0, 0]]
func = ot.PythonFunction(2, 1, f)
func.setDescription(["$s$", "$t$", "$cov$"])
cov_graph = func.draw([-a] * 2, [a] * 2, [512] * 2)
cov_graph.setLegendPosition("")
view = viewer.View(cov_graph)
plt.show()
Total running time of the script: (0 minutes 2.149 seconds)
 OpenTURNS
OpenTURNS