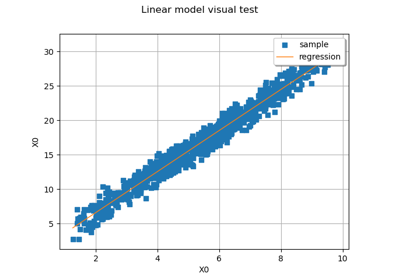
LinearModelAlgorithm¶
- class LinearModelAlgorithm(*args)¶
Class used to create a linear model from numerical samples.
- Parameters:
- XSample2-d sequence of float
The input samples of a model.
- YSample2-d sequence of float
The output samples of a model, must be of dimension 1.
- basis
Basis Optional. The
basis .
See also
Notes
This class is used in order to create a linear model from an input sample and an output sample. Let
be the sample size and let
be the input sample dimension. This class fits a linear regression model between the scalar variable
and the
-dimensional vector
. The linear model can be estimated with or without a functional basis.
If no basis is specified, the model is:
where
are unknown coefficients and
is a random variable with zero mean and constant (unknown) variance
independent from the coefficients
. The algorithm estimates the coefficients
of the linear model. Moreover, the method estimates the variance
.
If a functional basis is specified, let
be the number of functions in the basis. For
, let
be the
-th basis function. The linear model is:
where
is a random variable with zero mean and constant (and unknown) variance
and
are unknown coefficients. The algorithm estimates the coefficients
of the linear model. Moreover, the method estimates the variance
.
The coefficients
are evaluated using a linear least squares method, by default the QR method. User might also choose SVD or Cholesky by setting the LinearModelAlgorithm-DecompositionMethod key of the
ResourceMap. Here are a few guidelines to choose the appropriate decomposition method:The Cholesky can be safely used if the functional basis is orthogonal and the sample is drawn from the corresponding distribution, because this ensures that the columns of the design matrix are asymptotically orthogonal when the sample size increases. In this case, evaluating the Gram matrix does not increase the condition number.
Selecting the decomposition method can also be based on the sample size.
Please read the
Build()help page for details on this topic.The
LinearModelAnalysisclass can be used for a detailed analysis of the linear model result.No scaling is involved in this method. The scaling of the data, if any, is the responsibility of the user of the algorithm. This may be useful if, for example, we use a linear model (without functional basis) with very different input magnitudes and use the Cholesky decomposition applied to the associated Gram matrix. In this case, the Cholesky method may fail to produce accurate results.
Examples
>>> import openturns as ot >>> func = ot.SymbolicFunction( ... ['x1', 'x2', 'x3'], ... ['x1 + x2 + sin(x2 * 2 * pi_)/5 + 1e-3 * x3^2'] ... ) >>> dimension = 3 >>> distribution = ot.JointDistribution([ot.Normal()] * dimension) >>> inputSample = distribution.getSample(20) >>> outputSample = func(inputSample) >>> algo = ot.LinearModelAlgorithm(inputSample, outputSample) >>> algo.run() >>> result = algo.getResult() >>> design = result.getDesign() >>> gram = design.computeGram() >>> leverages = result.getLeverages()
In order to access the projection matrix, we build the least squares method.
>>> lsMethod = result.buildMethod() >>> projectionMatrix = lsMethod.getH()
Methods
BuildDistribution(inputSample)Recover the distribution, with metamodel performance in mind.
getBasis()Accessor to the input basis.
Accessor to the object's name.
Accessor to the joint probability density function of the physical input vector.
Accessor to the input sample.
getName()Accessor to the object's name.
Accessor to the output sample.
Accessor to the computed linear model.
Return the weights of the input sample.
hasName()Test if the object is named.
run()Compute the response surfaces.
setDistribution(distribution)Accessor to the joint probability density function of the physical input vector.
setName(name)Accessor to the object's name.
- __init__(*args)¶
- static BuildDistribution(inputSample)¶
Recover the distribution, with metamodel performance in mind.
For each marginal, find the best 1-d continuous parametric model else fallback to the use of a nonparametric one.
The selection is done as follow:
We start with a list of all parametric models (all factories)
For each model, we estimate its parameters if feasible.
We check then if model is valid, ie if its Kolmogorov score exceeds a threshold fixed in the MetaModelAlgorithm-PValueThreshold ResourceMap key. Default value is 5%
We sort all valid models and return the one with the optimal criterion.
For the last step, the criterion might be BIC, AIC or AICC. The specification of the criterion is done through the MetaModelAlgorithm-ModelSelectionCriterion ResourceMap key. Default value is fixed to BIC. Note that if there is no valid candidate, we estimate a non-parametric model (
KernelSmoothingorHistogram). The MetaModelAlgorithm-NonParametricModel ResourceMap key allows selecting the preferred one. Default value is HistogramOne each marginal is estimated, we use the Spearman independence test on each component pair to decide whether an independent copula. In case of non independence, we rely on a
NormalCopula.- Parameters:
- sample
Sample Input sample.
- sample
- Returns:
- distribution
Distribution Input distribution.
- distribution
- getBasis()¶
Accessor to the input basis.
- Returns:
- basis
Basis The basis which had been passed to the constructor.
- basis
- getClassName()¶
Accessor to the object’s name.
- Returns:
- class_namestr
The object class name (object.__class__.__name__).
- getDistribution()¶
Accessor to the joint probability density function of the physical input vector.
- Returns:
- distribution
Distribution Joint probability density function of the physical input vector.
- distribution
- getInputSample()¶
Accessor to the input sample.
- Returns:
- inputSample
Sample Input sample of a model evaluated apart.
- inputSample
- getName()¶
Accessor to the object’s name.
- Returns:
- namestr
The name of the object.
- getOutputSample()¶
Accessor to the output sample.
- Returns:
- outputSample
Sample Output sample of a model evaluated apart.
- outputSample
- getResult()¶
Accessor to the computed linear model.
- Returns:
- result
LinearModelResult The linear model built from numerical samples, along with other useful information.
- result
- getWeights()¶
Return the weights of the input sample.
- Returns:
- weightssequence of float
The weights of the points in the input sample.
- hasName()¶
Test if the object is named.
- Returns:
- hasNamebool
True if the name is not empty.
- run()¶
Compute the response surfaces.
Notes
It computes the response surfaces and creates a
MetaModelResultstructure containing all the results.
- setDistribution(distribution)¶
Accessor to the joint probability density function of the physical input vector.
- Parameters:
- distribution
Distribution Joint probability density function of the physical input vector.
- distribution
- setName(name)¶
Accessor to the object’s name.
- Parameters:
- namestr
The name of the object.
 OpenTURNS
OpenTURNS