Note
Go to the end to download the full example code.
Using otagrum¶
import openturns as ot
import pyagrum as gum
from matplotlib import pylab as plt
import otagrum
def showDot(dotstring):
try:
# fails outside notebook
import pyAgrum.lib.notebook as gnb
gnb.showDot(dotstring)
except ImportError:
import pydot
from io import BytesIO
graph = pydot.graph_from_dot_data(dotstring)[0]
with BytesIO() as f:
f.write(graph.create_png())
f.seek(0)
img = plt.imread(f)
fig = plt.imshow(img)
fig.axes.axis("off")
plt.show()
Creating the CBN structure We begin by creating the CBN that will be used throughout this example.
To do so, we need a NamedDAG structure…
dag = gum.DAG()
mapping = {}
mapping["A"] = dag.addNode() # Add node A
mapping["B"] = dag.addNode() # Add node B
mapping["C"] = dag.addNode() # Add node C
mapping["D"] = dag.addNode() # Add node D
dag.addArc(mapping["A"], mapping["C"]) # Arc A -> C
dag.addArc(mapping["B"], mapping["C"]) # Arc B -> C
dag.addArc(mapping["C"], mapping["D"]) # Arc C -> D
dag
(pyagrum.DAG@0x56237f0e96e0) {0,1,2,3} , {2->3,0->2,1->2}
structure = otagrum.NamedDAG(dag, list(mapping.keys()))
showDot(structure.toDot())
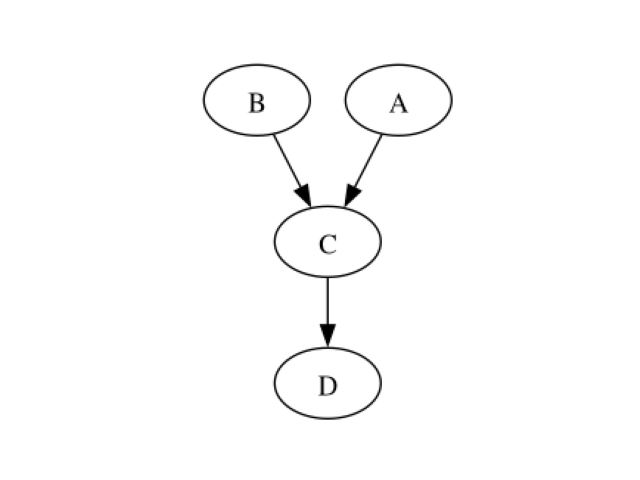
Parameters of the CBN … and a collection of marginals and local conditional copulas.
m_list = [ot.Uniform(0.0, 1.0) for i in range(structure.getSize())] # Local marginals
lcc_list = [] # Local Conditional Copulas
for i in range(structure.getSize()):
dim_lcc = structure.getParents(i).getSize() + 1
R = ot.CorrelationMatrix(dim_lcc)
for j in range(dim_lcc):
for k in range(j):
R[j, k] = 0.6
lcc_list.append(ot.Normal([0.0] * dim_lcc, [1.0] * dim_lcc, R).getCopula())
Now that we have a NamedDAG structure and a collection of local conditional copulas, we can construct a CBN.
cbn = otagrum.ContinuousBayesianNetwork(structure, m_list, lcc_list)
Having a CBN, we can now sample from it.
Learning the structure with continuous PC: Now that we have data, we can use it to learn the structure with the continuous PC algorithm.
learner = otagrum.ContinuousPC(sample, 5, 0.1)
We first learn the skeleton, that is the undirected structure.
skeleton = learner.learnSkeleton()
(pyagrum.UndiGraph@0x56237e8cc370) {0,1,2,3} , {2--3,0--2,1--2}
Then we look for the v-structures, leading to a Partially Directed Acyclic Graph (PDAG)
pdag = learner.learnPDAG()
(pyagrum.MixedGraph@0x56237ebc5ed0) {0,1,2,3} , {1->2,0->2} , {2--3}
Finally, the remaining edges are oriented by propagating constraints
ndag = learner.learnDAG()
showDot(ndag.toDot())
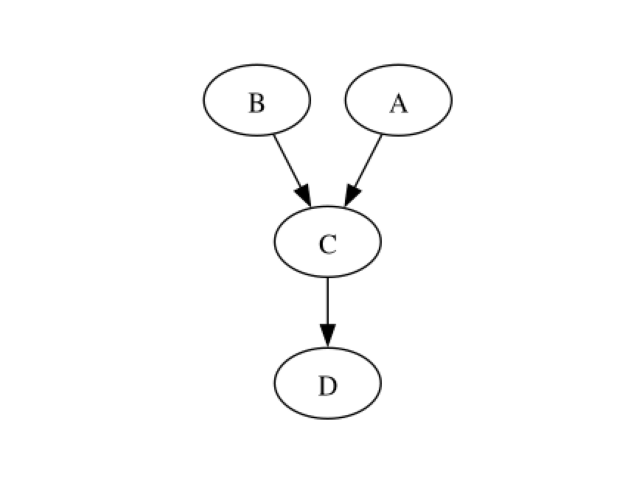
The true structure has been recovered. Learning with continuous MIIC Otagrum provides another learning algorithm to learn the structure: the continuous MIIC algorithm.
learner = otagrum.ContinuousMIIC(sample)
This algorithm relies on the computing of mutual information which is done through the copula function. Hence, a copula model for the data is needed. The continuous MIIC algorithm can make use of Gaussian copulas (parametric) or Bernstein copulas (non-parametric) to compute mutual information. Moreover, due to finite sampling size, the mutual information estimators need to be corrected. Two kind of correction are provided: NoCorr (no correction) or Naive (a fixed correction is subtracted from the raw mutual information estimators). Those behaviours can be changed as follows:
learner.setCMode(otagrum.CorrectedMutualInformation.CModeTypes_Bernstein) # By default
learner.setCMode(
otagrum.CorrectedMutualInformation.CModeTypes_Gaussian
) # To use Gaussian copulas
learner.setKMode(otagrum.CorrectedMutualInformation.KModeTypes_Naive) # By default
# learner.setKMode(otagrum.CorrectedMutualInformation.KModeTypes_NoCorr) # To use the raw estimators
learner.setAlpha(
0.01
) # Set the correction value for the Naive behaviour, it is set to 0.01 by default
As with PC algorithm we can learn the skeleton, PDAG and DAG using
skeleton = learner.learnSkeleton()
(pyagrum.UndiGraph@0x56237e859cc0) {0,1,2,3} , {2--3,0--2,1--2}
pdag = learner.learnPDAG()
(pyagrum.MixedGraph@0x56237f21b2e0) {0,1,2,3} , {2->3,1->2,0->2} , {}
dag = learner.learnDAG()
showDot(dag.toDot())
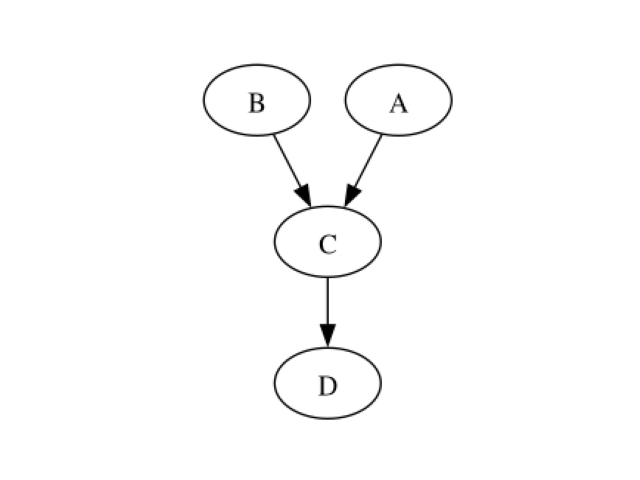
Learning parameters Bernstein copulas are used to learn the local conditional copulas associated to each node
m_list = []
lcc_list = []
for i in range(train.getDimension()):
m_list.append(ot.UniformFactory().build(train.getMarginal(i)))
indices = [i] + [int(n) for n in ndag.getParents(i)]
dim_lcc = len(indices)
if dim_lcc == 1:
bernsteinCopula = ot.IndependentCopula(1)
elif dim_lcc > 1:
K = otagrum.ContinuousTTest.GetK(len(train), dim_lcc)
bernsteinCopula = ot.EmpiricalBernsteinCopula(
train.getMarginal(indices), K, False
)
lcc_list.append(bernsteinCopula)
We can now create the learned CBN
lcbn = otagrum.ContinuousBayesianNetwork(ndag, m_list, lcc_list) # Learned CBN
And compare the mean loglikelihood between the true and the learned models
true_LL = compute_mean_LL(cbn, test)
print(true_LL)
0.31620906143480526
exp_LL = compute_mean_LL(lcbn, test)
print(exp_LL)
0.15241697100666607
Total running time of the script: (0 minutes 0.392 seconds)
 otagrum
otagrum