Create a linear model¶
In this example we are going to create a global approximation of a model response using linear model approximation.
Here
[1]:
from __future__ import print_function
import openturns as ot
try:
get_ipython()
except NameError:
import matplotlib
matplotlib.use('Agg')
from openturns.viewer import View
import numpy as np
import matplotlib.pyplot as plt
Hereafter we generate data using the previous model. We also add a noise:
[2]:
ot.RandomGenerator.SetSeed(0)
distribution = ot.Normal(2)
distribution.setDescription(["x","y"])
func = ot.SymbolicFunction(['x', 'y'], ['2 * x - y + 3 + 0.05 * sin(0.8*x)'])
input_sample = distribution.getSample(30)
epsilon = ot.Normal(0, 0.1).getSample(30)
output_sample = func(input_sample) + epsilon
Let us run the linear model algorithm using the LinearModelAlgorithm class & get its associated result :
[3]:
algo = ot.LinearModelAlgorithm(input_sample, output_sample)
result = ot.LinearModelResult(algo.getResult())
We get the result structure. As the underlying model is of type regression, it assumes a noise distribution associated to the residuals. Let us get it:
[4]:
print(result.getNoiseDistribution())
Normal(mu = 0, sigma = 0.110481)
We can get also residuals:
[5]:
print(result.getSampleResiduals())
[ y0 ]
0 : [ 0.186748 ]
1 : [ -0.117266 ]
2 : [ -0.039708 ]
3 : [ 0.10813 ]
4 : [ -0.0673202 ]
5 : [ -0.145401 ]
6 : [ 0.0184555 ]
7 : [ -0.103225 ]
8 : [ 0.107935 ]
9 : [ 0.0224636 ]
10 : [ -0.0432993 ]
11 : [ 0.0588534 ]
12 : [ 0.181832 ]
13 : [ 0.105051 ]
14 : [ -0.0433805 ]
15 : [ -0.175473 ]
16 : [ 0.211536 ]
17 : [ 0.0877925 ]
18 : [ -0.0367584 ]
19 : [ -0.0537763 ]
20 : [ -0.0838771 ]
21 : [ 0.0530871 ]
22 : [ 0.076703 ]
23 : [ -0.0940915 ]
24 : [ -0.0130962 ]
25 : [ 0.117419 ]
26 : [ -0.00233175 ]
27 : [ -0.0839944 ]
28 : [ -0.176839 ]
29 : [ -0.0561694 ]
We can get also standardized residuals (also called studentized residuals).
[6]:
print(result.getStandardizedResiduals())
0 : [ 1.80775 ]
1 : [ -1.10842 ]
2 : [ -0.402104 ]
3 : [ 1.03274 ]
4 : [ -0.633913 ]
5 : [ -1.34349 ]
6 : [ 0.198006 ]
7 : [ -0.980936 ]
8 : [ 1.01668 ]
9 : [ 0.20824 ]
10 : [ -0.400398 ]
11 : [ 0.563104 ]
12 : [ 1.68521 ]
13 : [ 1.02635 ]
14 : [ -0.406336 ]
15 : [ -1.63364 ]
16 : [ 2.07261 ]
17 : [ 0.85374 ]
18 : [ -0.342746 ]
19 : [ -0.498585 ]
20 : [ -0.781474 ]
21 : [ 0.497496 ]
22 : [ 0.759397 ]
23 : [ -0.930217 ]
24 : [ -0.121614 ]
25 : [ 1.11131 ]
26 : [ -0.0227866 ]
27 : [ -0.783004 ]
28 : [ -1.78814 ]
29 : [ -0.520003 ]
Now we got the result, we can perform a postprocessing analysis. We use LinearModelAnalysis for that purpose:
[7]:
analysis = ot.LinearModelAnalysis(result)
print(analysis)
Basis( [[x,y]->[1],[x,y]->[x],[x,y]->[y]] )
Coefficients:
| Estimate | Std Error | t value | Pr(>|t|) |
--------------------------------------------------------------------
[x,y]->[1] | 2.99847 | 0.0204173 | 146.859 | 9.82341e-41 |
[x,y]->[x] | 2.02079 | 0.0210897 | 95.8186 | 9.76973e-36 |
[x,y]->[y] | -0.994327 | 0.0215911 | -46.0527 | 3.35854e-27 |
--------------------------------------------------------------------
Residual standard error: 0.11048 on 27 degrees of freedom
F-statistic: 5566.3 , p-value: 0
---------------------------------
Multiple R-squared | 0.997581 |
Adjusted R-squared | 0.997401 |
---------------------------------
---------------------------------
Normality test | p-value |
---------------------------------
Anderson-Darling | 0.456553 |
Cramer-Von Mises | 0.367709 |
Chi-Squared | 0.669183 |
Kolmogorov-Smirnov | 0.578427 |
---------------------------------
It seems that the linear hypothesis could be accepted. Indeed, R-Squared value is nearly 1. Also the adjusted value (taking into account the datasize & number of parameters) is similar to R-Squared.
Also, we notice that both Fisher-Snedecor and Student p-values detailled above are less than 1%. This ensure the quality of the linear model.
Let us compare model and fitted values:
[8]:
analysis.drawModelVsFitted()
[8]:
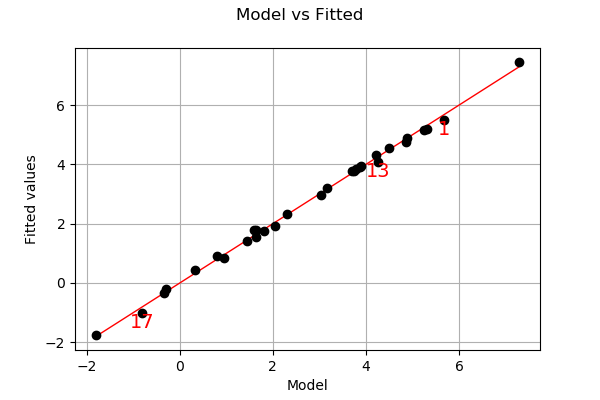
Seems that the linearity hypothesis is accurate.
We complete this analysis using some usefull graphs :
[9]:
fig = plt.figure(figsize=(12,10))
for k, plot in enumerate(["drawResidualsVsFitted", "drawScaleLocation", "drawQQplot",
"drawCookDistance", "drawResidualsVsLeverages", "drawCookVsLeverages"]):
graph = getattr(analysis, plot)()
ax = fig.add_subplot(3, 2, k + 1)
v = View(graph, figure=fig, axes=[ax])
_ = v.getFigure().suptitle("Diagnostic graphs", fontsize=18)
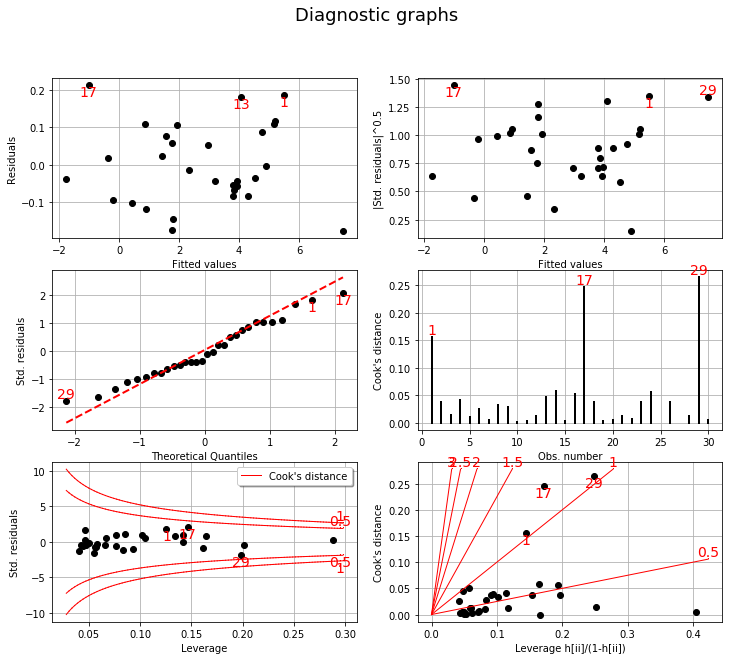
These graphics help asserting the linear model hypothesis. Indeed :
Quantile-to-quantile plot seems accurate
We notice heteroscedasticity within the noise
It seems that there is no outlier
Finally we give the intervals for each estimated coefficient (95% confidence interval):
[10]:
alpha = 0.95
interval = analysis.getCoefficientsConfidenceInterval(alpha)
print("confidence intervals with level=%1.2f : %s" % (alpha, interval))
confidence intervals with level=0.95 : [2.95657, 3.04036]
[1.97751, 2.06406]
[-1.03863, -0.950026]
 OpenTURNS
OpenTURNS