Note
Go to the end to download the full example code
Advanced kriging¶
In this example we will build a metamodel using gaussian process regression of the function.
We will choose the number of learning points, the basis and the covariance model.
import openturns as ot
from openturns.viewer import View
import numpy as np
import matplotlib.pyplot as plt
import openturns.viewer as viewer
ot.Log.Show(ot.Log.NONE)
Generate design of experiment¶
We create training samples from the function . We can change their number and distribution in the
range.
If the with_error boolean is True, then the data is computed by adding a gaussian noise to the function values.
dim = 1
xmin = 0
xmax = 10
n_pt = 20 # number of initial points
with_error = True # whether to use generation with error
ref_func_with_error = ot.SymbolicFunction(["x", "eps"], ["x * sin(x) + eps"])
ref_func = ot.ParametricFunction(ref_func_with_error, [1], [0.0])
x = np.vstack(np.linspace(xmin, xmax, n_pt))
ot.RandomGenerator.SetSeed(1235)
eps = ot.Normal(0, 1.5).getSample(n_pt)
X = ot.Sample(n_pt, 2)
X[:, 0] = x
X[:, 1] = eps
if with_error:
y = np.array(ref_func_with_error(X))
else:
y = np.array(ref_func(x))
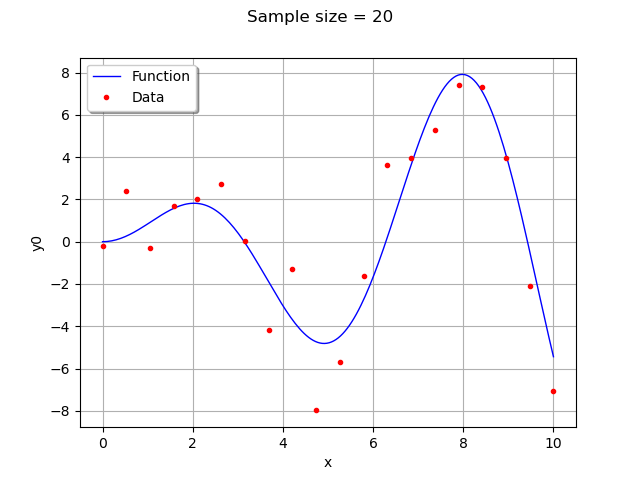
graph = ref_func.draw(xmin, xmax, 200)
cloud = ot.Cloud(x, y)
cloud.setColor("red")
cloud.setPointStyle("bullet")
graph.add(cloud)
graph.setLegends(["Function", "Data"])
graph.setLegendPosition("topleft")
graph.setTitle("Sample size = %d" % (n_pt))
view = viewer.View(graph)
Create the kriging algorithm¶
# 1. basis
ot.ResourceMap.SetAsBool(
"GeneralLinearModelAlgorithm-UseAnalyticalAmplitudeEstimate", True
)
basis = ot.ConstantBasisFactory(dim).build()
print(basis)
# 2. covariance model
cov = ot.MaternModel([1.0], [2.5], 1.5)
print(cov)
# 3. kriging algorithm
algokriging = ot.KrigingAlgorithm(x, y, cov, basis)
# error measure
# algokriging.setNoise([5*1e-1]*n_pt)
# 4. Optimization
# algokriging.setOptimizationAlgorithm(ot.NLopt('GN_DIRECT'))
lhsExperiment = ot.LHSExperiment(ot.Uniform(1e-1, 1e2), 50)
algokriging.setOptimizationAlgorithm(ot.MultiStart(ot.TNC(), lhsExperiment.generate()))
algokriging.setOptimizationBounds(ot.Interval([0.1], [1e2]))
# if we choose not to optimize parameters
# algokriging.setOptimizeParameters(False)
# 5. run the algorithm
algokriging.run()
Basis( [class=LinearEvaluation name=Unnamed center=[0] constant=[1] linear=[[ 0 ]]] )
MaternModel(scale=[1], amplitude=[2.5], nu=1.5)
Results¶
get some results
krigingResult = algokriging.getResult()
print("residual = ", krigingResult.getResiduals())
print("R2 = ", krigingResult.getRelativeErrors())
print("Optimal scale= {}".format(krigingResult.getCovarianceModel().getScale()))
print(
"Optimal amplitude = {}".format(krigingResult.getCovarianceModel().getAmplitude())
)
print("Optimal trend coefficients = {}".format(krigingResult.getTrendCoefficients()))
residual = [0.00140221]
R2 = [2.02353e-06]
Optimal scale= [0.821466]
Optimal amplitude = [4.51497]
Optimal trend coefficients = [[-0.118004]]
get the metamodel
krigingMeta = krigingResult.getMetaModel()
n_pts_plot = 1000
x_plot = np.vstack(np.linspace(xmin, xmax, n_pts_plot))
fig, [ax1, ax2] = plt.subplots(1, 2, figsize=(12, 6))
# On the left, the function
graph = ref_func.draw(xmin, xmax, n_pts_plot)
graph.setLegends(["Function"])
graphKriging = krigingMeta.draw(xmin, xmax, n_pts_plot)
graphKriging.setColors(["green"])
graphKriging.setLegends(["Kriging"])
graph.add(graphKriging)
cloud = ot.Cloud(x, y)
cloud.setColor("red")
cloud.setLegend("Data")
graph.add(cloud)
graph.setLegendPosition("topleft")
View(graph, axes=[ax1])
# On the right, the conditional kriging variance
graph = ot.Graph("", "x", "Conditional kriging variance", True, "")
# Sample for the data
sample = ot.Sample(n_pt, 2)
sample[:, 0] = x
cloud = ot.Cloud(sample)
cloud.setColor("red")
graph.add(cloud)
# Sample for the variance
sample = ot.Sample(n_pts_plot, 2)
sample[:, 0] = x_plot
variance = [[krigingResult.getConditionalCovariance(xx)[0, 0]] for xx in x_plot]
sample[:, 1] = variance
curve = ot.Curve(sample)
curve.setColor("green")
graph.add(curve)
View(graph, axes=[ax2])
fig.suptitle("Kriging result")
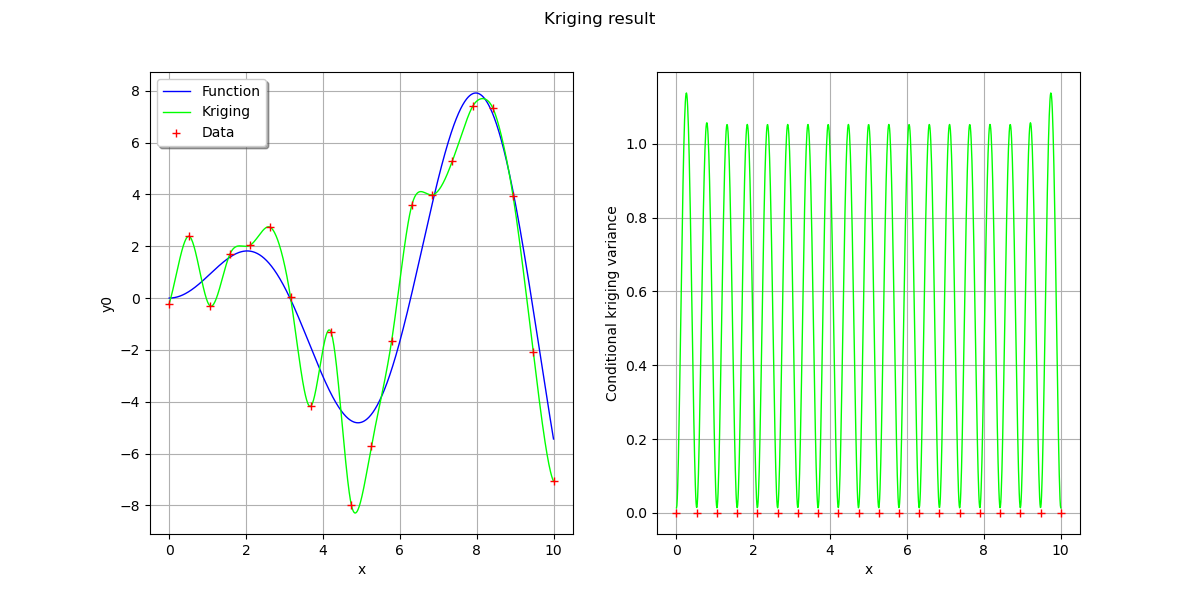
Text(0.5, 0.98, 'Kriging result')
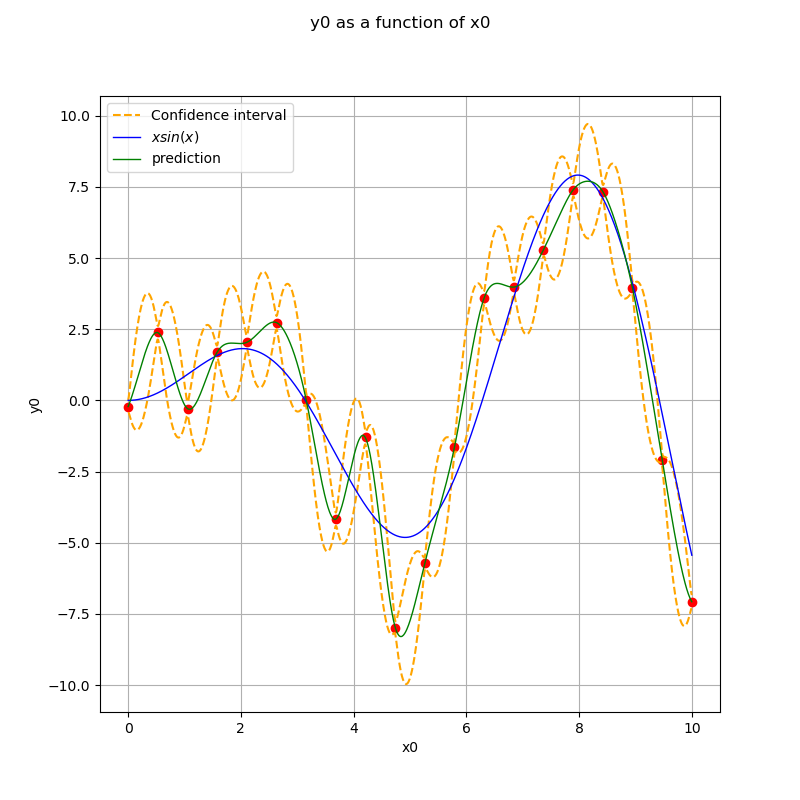
Display the confidence interval¶
sphinx_gallery_thumbnail_number = 3
level = 0.95
quantile = ot.Normal().computeQuantile((1 - level) / 2)[0]
borne_sup = krigingMeta(x_plot) + quantile * np.sqrt(variance)
borne_inf = krigingMeta(x_plot) - quantile * np.sqrt(variance)
fig, ax = plt.subplots(figsize=(8, 8))
ax.plot(x, y, ("ro"))
ax.plot(x_plot, borne_sup, "--", color="orange", label="Confidence interval")
ax.plot(x_plot, borne_inf, "--", color="orange")
View(ref_func.draw(xmin, xmax, n_pts_plot), axes=[ax], plot_kw={"label": "$x sin(x)$"})
View(
krigingMeta.draw(xmin, xmax, n_pts_plot),
plot_kw={"color": "green", "label": "prediction"},
axes=[ax],
)
legend = ax.legend()
ax.autoscale()
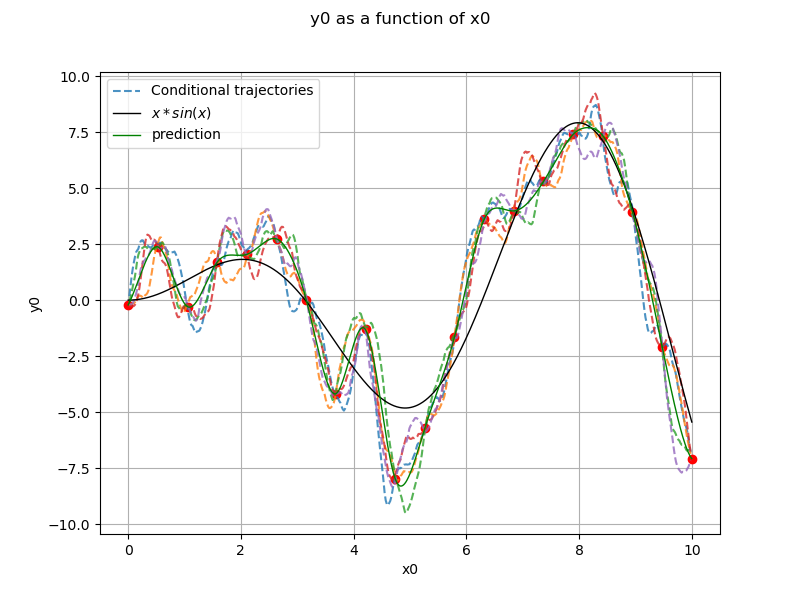
Generate conditional trajectories¶
support for trajectories with training samples removed
values = np.linspace(0, 10, 500)
for xx in x:
if len(np.argwhere(values == xx)) == 1:
values = np.delete(values, np.argwhere(values == xx)[0, 0])
Conditional Gaussian process
krv = ot.KrigingRandomVector(krigingResult, np.vstack(values))
krv_sample = krv.getSample(5)
x_plot = np.vstack(np.linspace(xmin, xmax, n_pts_plot))
fig, ax = plt.subplots(figsize=(8, 6))
ax.plot(x, y, ("ro"))
for i in range(krv_sample.getSize()):
if i == 0:
ax.plot(
values, krv_sample[i, :], "--", alpha=0.8, label="Conditional trajectories"
)
else:
ax.plot(values, krv_sample[i, :], "--", alpha=0.8)
View(
ref_func.draw(xmin, xmax, n_pts_plot),
axes=[ax],
plot_kw={"color": "black", "label": "$x*sin(x)$"},
)
View(
krigingMeta.draw(xmin, xmax, n_pts_plot),
axes=[ax],
plot_kw={"color": "green", "label": "prediction"},
)
legend = ax.legend()
ax.autoscale()
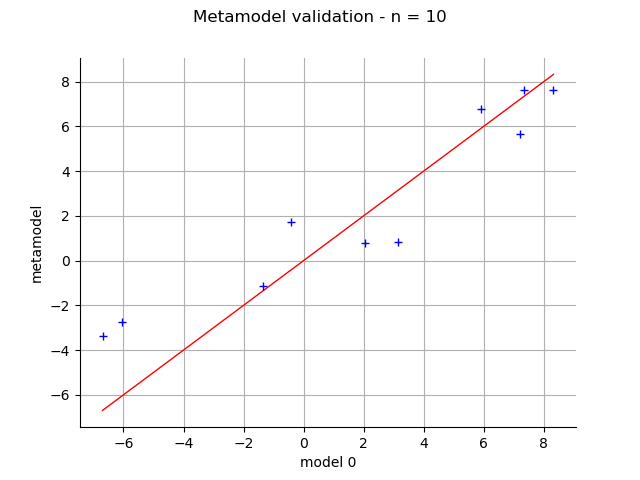
Validation¶
n_valid = 10
x_valid = ot.Uniform(xmin, xmax).getSample(n_valid)
if with_error:
X_valid = ot.Sample(x_valid)
X_valid.stack(ot.Normal(0.0, 1.5).getSample(n_valid))
y_valid = np.array(ref_func_with_error(X_valid))
else:
y_valid = np.array(ref_func(X_valid))
validation = ot.MetaModelValidation(x_valid, y_valid, krigingMeta)
validation.computePredictivityFactor()
graph = validation.drawValidation()
view = viewer.View(graph)
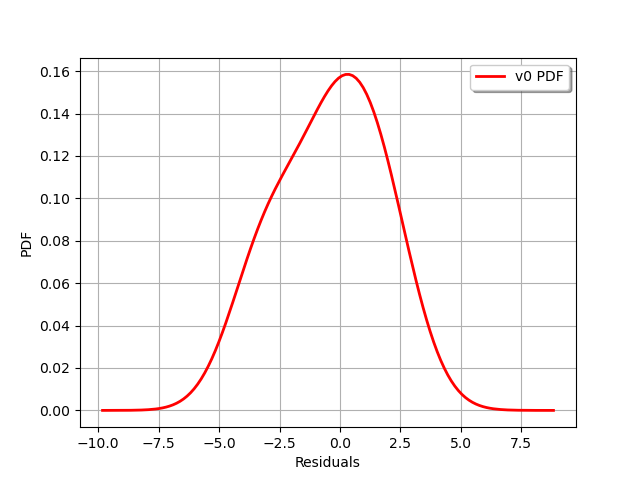
graph = validation.getResidualDistribution().drawPDF()
graph.setXTitle("Residuals")
view = viewer.View(graph)
plt.show()
Total running time of the script: ( 0 minutes 0.768 seconds)
 OpenTURNS
OpenTURNS