Note
Go to the end to download the full example code
Use the ANCOVA indices¶
In this example we are going to use the ANCOVA decomposition to estimate sensitivity indices from a model with correlated inputs.
ANCOVA allows one to estimate the Sobol’ indices, and thanks to a functional decomposition of the model it allows one to separate the part of variance explained by a variable itself from the part of variance explained by a correlation which is due to its correlation with the other input parameters.
In theory, ANCOVA indices range is ; the closer to 1 the index is,
the greater the model response sensitivity to the variable is.
These indices are a sum of a physical part
and correlated part
.
The correlation has a weak influence on the contribution of
, if
is low and
is close to
.
On the contrary, the correlation has a strong influence on the contribution of
the input
, if
is high and
is close to
.
The ANCOVA indices evaluate the importance of one variable at a time
(
indices stored, with
the input dimension of the model).
The
uncorrelated parts of variance of the output due to each input
and the effects of the correlation are represented by the indices resulting
from the subtraction of these two previous lists.
To evaluate the indices, the library needs of a functional chaos result approximating the model response with uncorrelated inputs and a sample with correlated inputs used to compute the real values of the output.
import openturns as ot
import openturns.viewer as viewer
from matplotlib import pylab as plt
ot.Log.Show(ot.Log.NONE)
Create the model (x1,x2) –> (y) = (4.*x1+5.*x2)
model = ot.SymbolicFunction(["x1", "x2"], ["4.*x1+5.*x2"])
Create the input independent joint distribution
distribution = ot.Normal(2)
distribution.setDescription(["X1", "X2"])
Create the correlated input distribution
S = ot.CorrelationMatrix(2)
S[1, 0] = 0.3
R = ot.NormalCopula.GetCorrelationFromSpearmanCorrelation(S)
copula = ot.NormalCopula(R)
distribution_corr = ot.ComposedDistribution([ot.Normal()] * 2, copula)
ANCOVA needs a functional decomposition of the model
enumerateFunction = ot.LinearEnumerateFunction(2)
productBasis = ot.OrthogonalProductPolynomialFactory(
[ot.HermiteFactory()] * 2, enumerateFunction
)
adaptiveStrategy = ot.FixedStrategy(
productBasis, enumerateFunction.getStrataCumulatedCardinal(4)
)
samplingSize = 250
experiment = ot.MonteCarloExperiment(distribution, samplingSize)
X = experiment.generate()
Y = model(X)
algo = ot.FunctionalChaosAlgorithm(X, Y, distribution, adaptiveStrategy)
algo.run()
result = ot.FunctionalChaosResult(algo.getResult())
Create the input sample taking account the correlation
size = 2000
sample = distribution_corr.getSample(size)
Perform the decomposition
ancova = ot.ANCOVA(result, sample)
# Compute the ANCOVA indices (first order and uncorrelated indices are computed together)
indices = ancova.getIndices()
# Retrieve uncorrelated indices
uncorrelatedIndices = ancova.getUncorrelatedIndices()
# Retrieve correlated indices:
correlatedIndices = indices - uncorrelatedIndices
Print Sobol’ indices, the physical part, and the correlation part
print("ANCOVA indices ", indices)
print("ANCOVA uncorrelated indices ", uncorrelatedIndices)
print("ANCOVA correlated indices ", correlatedIndices)
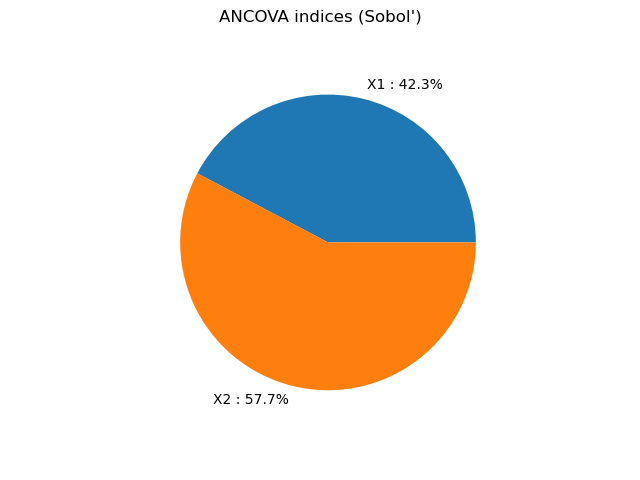
ANCOVA indices [0.422633,0.577367]
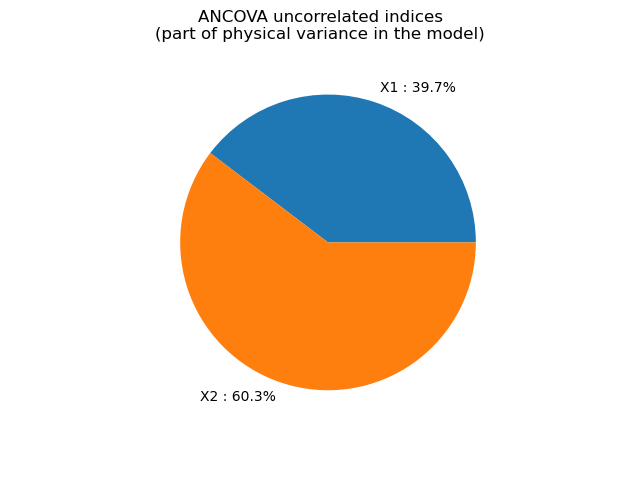
ANCOVA uncorrelated indices [0.296946,0.451679]
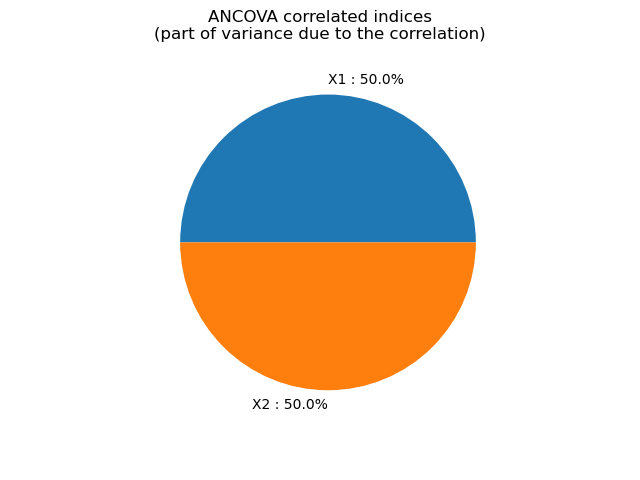
ANCOVA correlated indices [0.125687,0.125687]
graph = ot.SobolIndicesAlgorithm.DrawImportanceFactors(
indices, distribution.getDescription(), "ANCOVA indices (Sobol')"
)
view = viewer.View(graph)
graph = ot.SobolIndicesAlgorithm.DrawImportanceFactors(
uncorrelatedIndices,
distribution.getDescription(),
"ANCOVA uncorrelated indices\n(part of physical variance in the model)",
)
view = viewer.View(graph)
graph = ot.SobolIndicesAlgorithm.DrawImportanceFactors(
correlatedIndices,
distribution.getDescription(),
"ANCOVA correlated indices\n(part of variance due to the correlation)",
)
view = viewer.View(graph)
plt.show()
Total running time of the script: ( 0 minutes 0.137 seconds)
 OpenTURNS
OpenTURNS