Note
Go to the end to download the full example code
Various design of experiments in OpenTURNS¶
The goal of this example is to present several design of experiments available in OpenTURNS.
Distribution¶
import openturns as ot
import openturns.viewer as otv
ot.Log.Show(ot.Log.NONE)
Monte-Carlo sampling in 2D¶
dim = 2
X = [ot.Uniform()] * dim
distribution = ot.ComposedDistribution(X)
bounds = distribution.getRange()
sampleSize = 10
sample = distribution.getSample(sampleSize)
fig = otv.PlotDesign(sample, bounds)
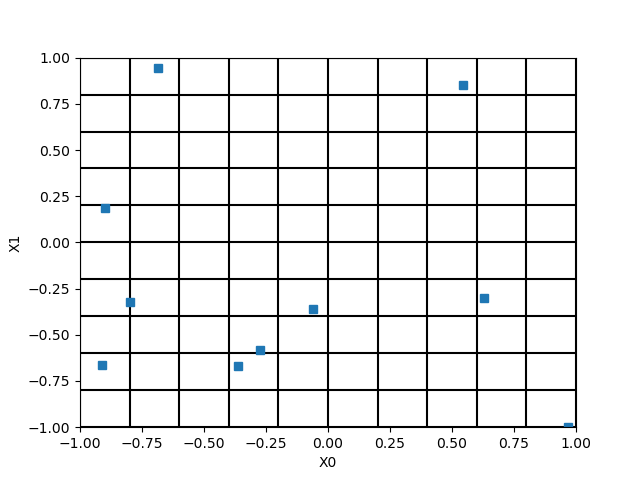
We see that there a empty zones in the input space.
Monte-Carlo sampling in 3D¶
dim = 3
X = [ot.Uniform()] * dim
distribution = ot.ComposedDistribution(X)
bounds = distribution.getRange()
sampleSize = 10
sample = distribution.getSample(sampleSize)
fig = otv.PlotDesign(sample, bounds)
fig.set_size_inches(10, 10)
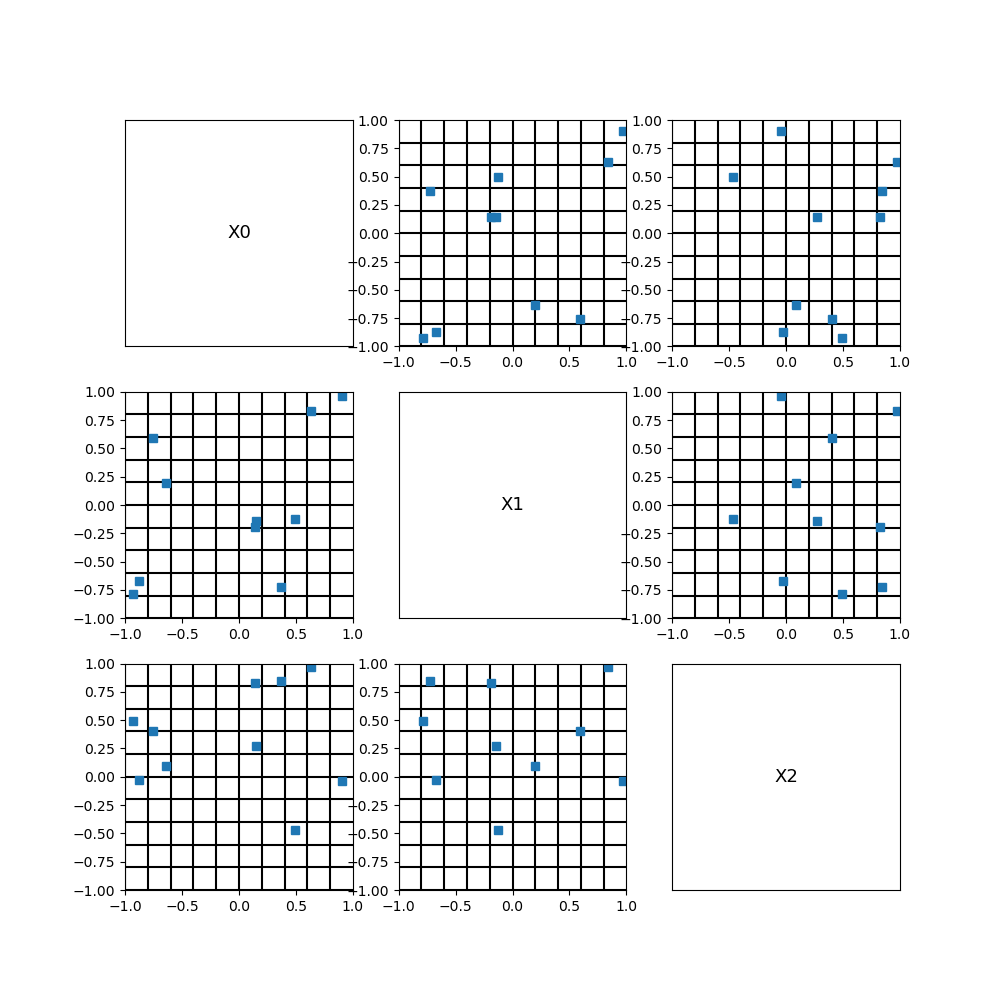
Latin Hypercube Sampling¶
distribution = ot.ComposedDistribution([ot.Uniform()] * 3)
samplesize = 5
experiment = ot.LHSExperiment(distribution, samplesize, False, False)
sample = experiment.generate()
In order to see the LHS property, we need to set the bounds.
bounds = distribution.getRange()
fig = otv.PlotDesign(sample, bounds)
fig.set_size_inches(10, 10)
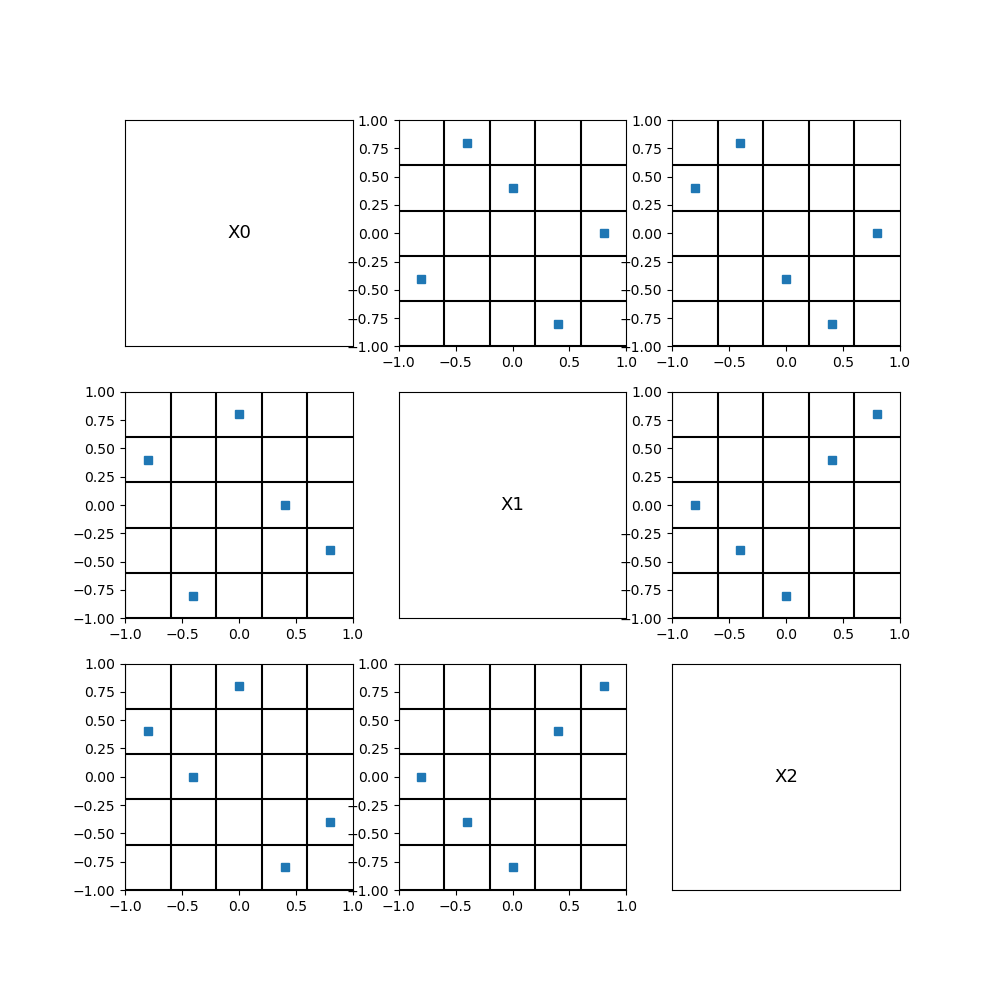
We see that each column or row exactly contains one single point. This shows that a LHS design of experiments has good 1D projection properties, and, hence, is a good candidate for a space filling design.
Optimized LHS¶
distribution = ot.ComposedDistribution([ot.Uniform()] * 3)
samplesize = 10
bounds = distribution.getRange()
lhs = ot.LHSExperiment(distribution, samplesize)
lhs.setAlwaysShuffle(True) # randomized
space_filling = ot.SpaceFillingC2()
temperatureProfile = ot.GeometricProfile(10.0, 0.95, 1000)
algo = ot.SimulatedAnnealingLHS(lhs, space_filling, temperatureProfile)
# optimal design
sample = algo.generate()
fig = otv.PlotDesign(sample, bounds)
fig.set_size_inches(10, 10)
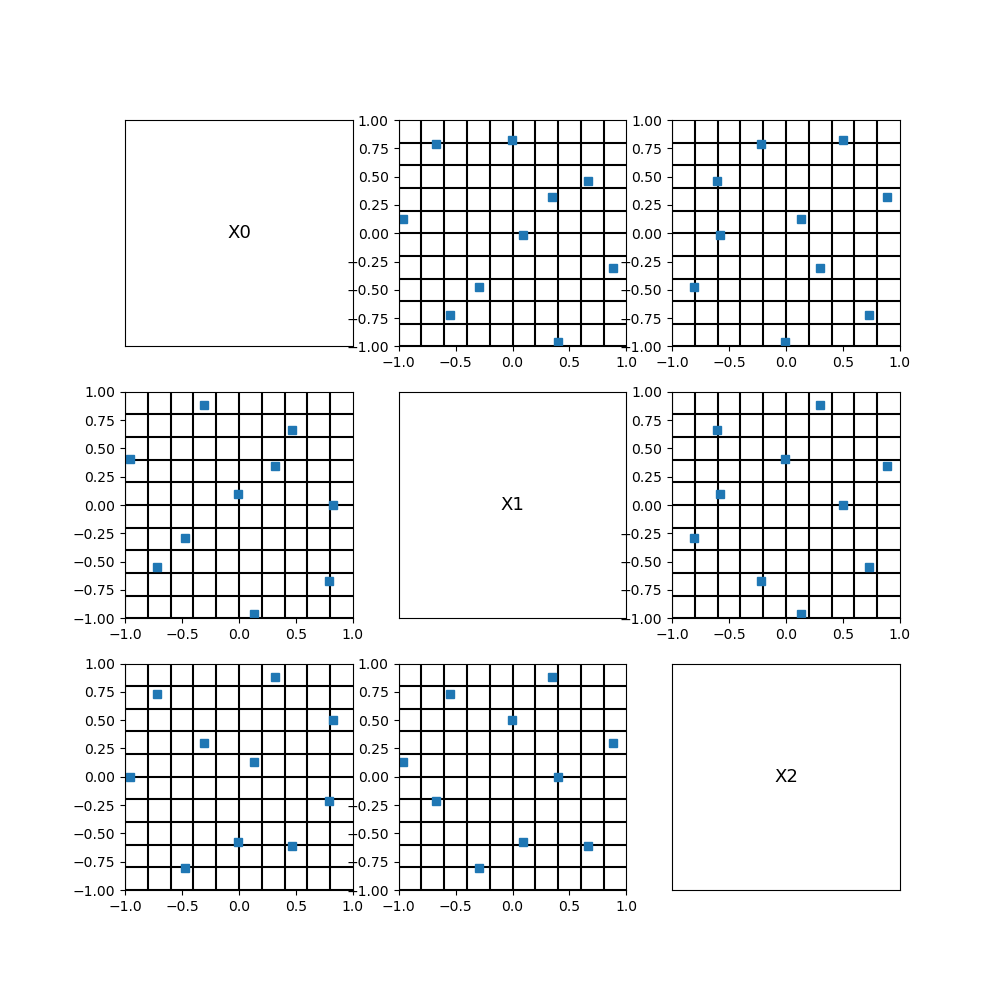
We see that this LHS is optimized in the sense that it fills the space more evenly than a non-optimized does in general.
Sobol’ low discrepancy sequence¶
dim = 2
distribution = ot.ComposedDistribution([ot.Uniform()] * dim)
bounds = distribution.getRange()
sequence = ot.SobolSequence(dim)
samplesize = 2 ** 5 # Sobol' sequences are in base 2
experiment = ot.LowDiscrepancyExperiment(sequence, distribution, samplesize, False)
sample = experiment.generate()
samplesize
32
subdivisions = [2 ** 2, 2 ** 1]
fig = otv.PlotDesign(sample, bounds, subdivisions)
fig.set_size_inches(6, 6)
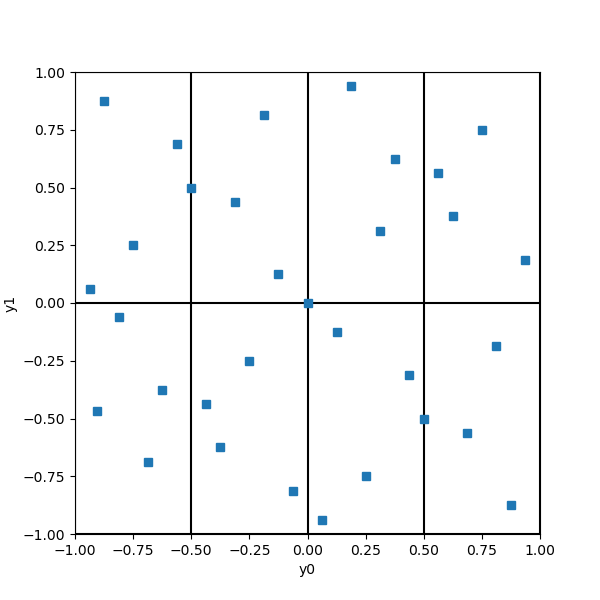
We have elementary intervals in 2 dimensions, each having a volume equal to 1/8. Since there are 32 points, the Sobol’ sequence is so that each elementary interval contains exactly 32/8 = 4 points. Notice that each elementary interval is closed on the left (or bottom) and open on the right (or top).
Halton low discrepancy sequence¶
dim = 2
distribution = ot.ComposedDistribution([ot.Uniform()] * dim)
bounds = distribution.getRange()
sequence = ot.HaltonSequence(dim)
Halton sequence uses prime numbers 2 and 3 in two dimensions.
samplesize = 2 ** 2 * 3 ** 2
experiment = ot.LowDiscrepancyExperiment(sequence, distribution, samplesize, False)
sample = experiment.generate()
samplesize
36
subdivisions = [2 ** 2, 3]
fig = otv.PlotDesign(sample, bounds, subdivisions)
fig.set_size_inches(6, 6)
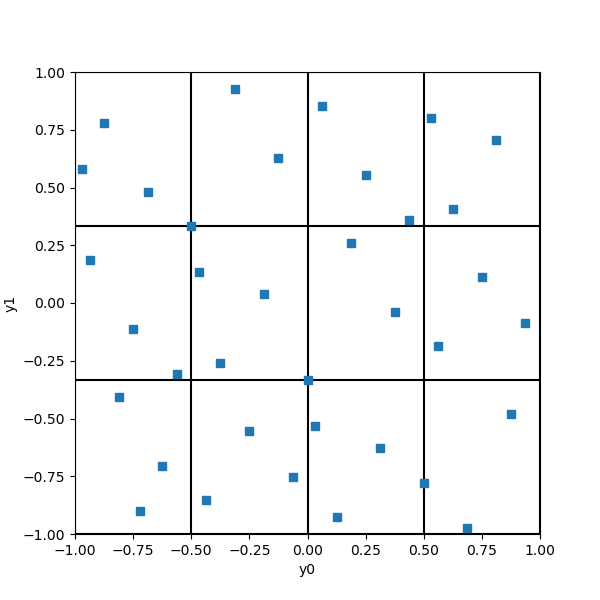
We have elementary intervals in 2 dimensions, each having a volume equal to 1/12. Since there are 36 points, the Halton sequence is so that each elementary interval contains exactly 36/12 = 3 points. Notice that each elementary interval is closed on the left (or bottom) and open on the right (or top).
 OpenTURNS
OpenTURNS