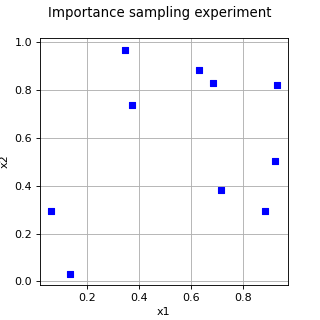
ImportanceSamplingExperiment¶
(Source code, png)
- class ImportanceSamplingExperiment(*args)¶
Importance Sampling experiment.
- Available constructors:
ImportanceSamplingExperiment(importanceDistribution)
ImportanceSamplingExperiment(importanceDistribution, size)
ImportanceSamplingExperiment(initialDistribution, importanceDistribution, size)
- Parameters:
- initialDistribution
Distribution Distribution
which is the initial distribution used to generate the set of input data.
- sizepositive int
Number of points that will be generated in the experiment.
- importanceDistribution
Distribution Distribution
according to which the points of the experiments will be generated with the Importance Sampling technique.
- initialDistribution
See also
Notes
ImportanceSamplingExperiment is a random weighted design of experiments. We can use it to generate a sample
based on independent observations from the distribution
(see [hammersley1961] page 57, [lemieux2009] page 11). Importance sampling is a variance reduction method i.e. it aims at reducing the variance of the estimator of the weighted integral. The sample is generated from the importance distribution
and each realization is weighted by
where
is the probability density function of the input random vector.
There is no general method in the library to provide the importance distribution, which must be specified by the user. In the specific case of rare event estimation, [morio2015] page 53 provides different methods to compute the instrumental distribution, including simple changes of measure and exponential twisting. The
PostAnalyticalImportanceSamplingclass combines theFORMclass and importance sampling to estimate a probability.The ImportanceSamplingExperiment class is nonadaptive, i.e. the parameters of the instrumental distribution are set once for all. See
NAISfor an adaptive importance sampling algorithm andCrossEntropyImportanceSamplingfor another algorithm.Examples
>>> import openturns as ot >>> ot.RandomGenerator.SetSeed(0) >>> distribution = ot.JointDistribution([ot.Uniform(0, 1)] * 2) >>> importanceDistribution = ot.JointDistribution([ot.Uniform(0, 1)] * 2) >>> experiment = ot.ImportanceSamplingExperiment(distribution, importanceDistribution, 5) >>> print(experiment.generate()) [ X0 X1 ] 0 : [ 0.629877 0.882805 ] 1 : [ 0.135276 0.0325028 ] 2 : [ 0.347057 0.969423 ] 3 : [ 0.92068 0.50304 ] 4 : [ 0.0632061 0.292757 ]
Methods
generate()Generate points according to the type of the experiment.
Generate points and their associated weight according to the type of the experiment.
Accessor to the object's name.
Accessor to the distribution.
Accessor to the importance distribution.
getName()Accessor to the object's name.
getSize()Accessor to the size of the generated sample.
hasName()Test if the object is named.
Ask whether the experiment has uniform weights.
isRandom()Accessor to the randomness of quadrature.
setDistribution(distribution)Accessor to the distribution.
setName(name)Accessor to the object's name.
setSize(size)Accessor to the size of the generated sample.
- __init__(*args)¶
- generate()¶
Generate points according to the type of the experiment.
- Returns:
- sample
Sample Points
of the design of experiments. The sampling method is defined by the type of the weighted experiment.
- sample
Examples
>>> import openturns as ot >>> ot.RandomGenerator.SetSeed(0) >>> myExperiment = ot.MonteCarloExperiment(ot.Normal(2), 5) >>> sample = myExperiment.generate() >>> print(sample) [ X0 X1 ] 0 : [ 0.608202 -1.26617 ] 1 : [ -0.438266 1.20548 ] 2 : [ -2.18139 0.350042 ] 3 : [ -0.355007 1.43725 ] 4 : [ 0.810668 0.793156 ]
- generateWithWeights()¶
Generate points and their associated weight according to the type of the experiment.
- Returns:
Examples
>>> import openturns as ot >>> ot.RandomGenerator.SetSeed(0) >>> myExperiment = ot.MonteCarloExperiment(ot.Normal(2), 5) >>> sample, weights = myExperiment.generateWithWeights() >>> print(sample) [ X0 X1 ] 0 : [ 0.608202 -1.26617 ] 1 : [ -0.438266 1.20548 ] 2 : [ -2.18139 0.350042 ] 3 : [ -0.355007 1.43725 ] 4 : [ 0.810668 0.793156 ] >>> print(weights) [0.2,0.2,0.2,0.2,0.2]
- getClassName()¶
Accessor to the object’s name.
- Returns:
- class_namestr
The object class name (object.__class__.__name__).
- getDistribution()¶
Accessor to the distribution.
- Returns:
- distribution
Distribution Distribution of the input random vector.
- distribution
- getImportanceDistribution()¶
Accessor to the importance distribution.
- Returns:
- importanceDistribution
Distribution Distribution
according to which the points of the design of experiments will be generated with the Importance Sampling technique.
- importanceDistribution
- getName()¶
Accessor to the object’s name.
- Returns:
- namestr
The name of the object.
- getSize()¶
Accessor to the size of the generated sample.
- Returns:
- sizepositive int
Number
of points constituting the design of experiments.
- hasName()¶
Test if the object is named.
- Returns:
- hasNamebool
True if the name is not empty.
- hasUniformWeights()¶
Ask whether the experiment has uniform weights.
- Returns:
- hasUniformWeightsbool
Whether the experiment has uniform weights.
- isRandom()¶
Accessor to the randomness of quadrature.
- Parameters:
- isRandombool
Is true if the design of experiments is random. Otherwise, the design of experiment is assumed to be deterministic.
- setDistribution(distribution)¶
Accessor to the distribution.
- Parameters:
- distribution
Distribution Distribution of the input random vector.
- distribution
- setName(name)¶
Accessor to the object’s name.
- Parameters:
- namestr
The name of the object.
- setSize(size)¶
Accessor to the size of the generated sample.
- Parameters:
- sizepositive int
Number
of points constituting the design of experiments.
Examples using the class¶
Axial stressed beam : comparing different methods to estimate a probability
 OpenTURNS
OpenTURNS