BernsteinCopulaFactory¶
- class BernsteinCopulaFactory(*args)¶
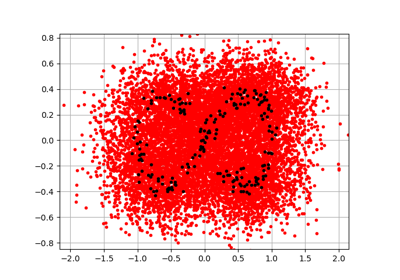
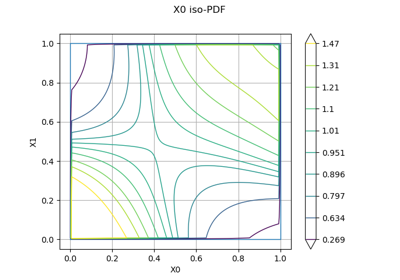
EmpiricalBernsteinCopula factory.
Methods
ComputeAMISEBinNumber(sample)Compute the optimal AMISE number of bins.
Compute the optimal log-likelihood number of bins by cross-validation.
Compute the optimal penalized Csiszar divergence number of bins.
build(*args)Build the empirical Bernstein copula.
Build the empirical Bernstein copula as a native distribution.
buildEstimator(*args)Build the distribution and the parameter distribution.
Accessor to the bootstrap size.
Accessor to the object's name.
Accessor to the known parameters indices.
Accessor to the known parameters values.
getName()Accessor to the object's name.
hasName()Test if the object is named.
setBootstrapSize(bootstrapSize)Accessor to the bootstrap size.
setKnownParameter(values, positions)Accessor to the known parameters.
setName(name)Accessor to the object's name.
See also
Notes
This class builds an
EmpiricalBernsteinCopulawhich is a non parametric fitting of the copula of a multivariate distribution.The keys of
ResourceMaprelated to the class are:the keys BernsteinCopulaFactory-MinM and BernsteinCopulaFactory-MaxM that define the range of
in the optimization problems computing the optimal bin number according to a specified criterion,
the key BernsteinCopulaFactory-BinNumberSelection that defines the criterion to compute the optimal bin number when it is not specified. The possible choices are ‘AMISE’, ‘LogLikelihood’, ‘PenalizedCsiszarDivergence’;
the key BernsteinCopulaFactory-KFraction that defines the fraction of the sample used for the validation in the method
ComputeLogLikelihoodBinNumber(),the key BernsteinCopulaFactory-SamplingSize that defines the
parameter used in the method
ComputePenalizedCsiszarDivergenceBinNumber().
- __init__(*args)¶
- static ComputeAMISEBinNumber(sample)¶
Compute the optimal AMISE number of bins.
- Parameters:
- sample2-d sequence of float, of dimension 1
The sample from which the optimal AMISE bin number is computed.
Notes
The bin number
is computed by minimizing the asymptotic mean integrated squared error (AMISE), leading to:
where
is the largest integer less than or equal to
,
the sample size and
the sample dimension.
Note that this optimal
does not necessarily divide the sample size
.
- static ComputeLogLikelihoodBinNumber(*args)¶
Compute the optimal log-likelihood number of bins by cross-validation.
- Parameters:
- sample2-d sequence of float, of dimension 1
The sample of size
from which the optimal log-likelihood bin number is computed.
- kFractionint,
The fraction of the sample used for the validation.
Default value 2.
Notes
Let
be the given sample. If
, the bin number
is given by:
where
is the density function of the
EmpiricalBernsteinCopulaassociated to the sampleand the bin number
.
If
, the bin number
is given by:
where
and
.
Note that this optimal
does not necessarily divide the sample size
.
- static ComputePenalizedCsiszarDivergenceBinNumber(*args)¶
Compute the optimal penalized Csiszar divergence number of bins.
- Parameters:
- sample2-d sequence of float, of dimension 1
The sample of size
from which the optimal AMISE bin number is computed.
- f
Function The function defining the Csiszar divergence of interest.
- alphafloat,
The penalization factor.
Notes
Let
be the given sample. The bin number
is given by:
where
is the density function of the
EmpiricalBernsteinCopulaassociated to the sampleand the bin number
,
a Monte Carlo estimate of the Csiszar
divergence,
the exact Spearman correlation of the empirical Bernstein copula
and
the empirical Spearman correlation of the sample
.
The parameter
is controlled by the BernsteinCopulaFactory-SamplingSize key in
ResourceMap.Note that this optimal
does not necessarily divide the sample size
.
- build(*args)¶
Build the empirical Bernstein copula.
Available usages:
build()
build(sample)
build(sample, m)
build(sample, method, f)
- Parameters:
- sample2-d sequence of float, of dimension
The sample of size
from which the copula is estimated.
- methodstr
The name of the bin number selection method. Possible choices are AMISE, LogLikelihood and PenalizedCsiszarDivergence.
Default is LogLikelihood.
- f
Function The function defining the Csiszar divergence of interest used by the PenalizedCsiszarDivergence method.
Default is Function().
- mint,:math:1 leq m leq sampleSize,
The bin number, i.e. the number of sub-intervals in which all the edges of the unit cube
are regularly partitioned.
Default value is the value computed from the default bin number selection method.
- sample2-d sequence of float, of dimension
- Returns:
- copula
Distribution The empirical Bernstein copula as a generic distribution.
- copula
Notes
If the bin number
is specified and does not divide the sample size
, then a part of the sample is removed for the result to be a copula. See
EmpiricalBernsteinCopula.
- buildAsEmpiricalBernsteinCopula(*args)¶
Build the empirical Bernstein copula as a native distribution.
Available usages:
buildAsEmpiricalBernsteinCopula()
buildAsEmpiricalBernsteinCopula(sample)
buildAsEmpiricalBernsteinCopula(sample, m)
buildAsEmpiricalBernsteinCopula(sample, method, f)
- Parameters:
- sample2-d sequence of float, of dimension d
The sample of size
from which the copula is estimated.
- methodstr
The name of the bin number selection method. Possible choices are AMISE, LogLikelihood and PenalizedCsiszarDivergence.
Default is LogLikelihood.
- f
Function The function defining the Csiszar divergence of interest used by the PenalizedCsiszarDivergence method.
Default is Function().
- mint,
,
The bin number, i.e. the number of sub-intervals in which all the edges of the unit cube
are regularly partitioned.
Default value is the value computed from the default bin number selection method.
- Returns:
- copula
EmpiricalBernsteinCopula The empirical Bernstein copula as a native distribution.
- copula
Notes
If the bin number
is specified and does not divide the sample size
, then a part of the sample is removed for the result to be a copula a copula. See
EmpiricalBernsteinCopula.
- buildEstimator(*args)¶
Build the distribution and the parameter distribution.
- Parameters:
- sample2-d sequence of float
Data.
- parameters
DistributionParameters Optional, the parametrization.
- Returns:
- resDist
DistributionFactoryResult The results.
- resDist
Notes
According to the way the native parameters of the distribution are estimated, the parameters distribution differs:
Moments method: the asymptotic parameters distribution is normal and estimated by Bootstrap on the initial data;
Maximum likelihood method with a regular model: the asymptotic parameters distribution is normal and its covariance matrix is the inverse Fisher information matrix;
Other methods: the asymptotic parameters distribution is estimated by Bootstrap on the initial data and kernel fitting (see
KernelSmoothing).
If another set of parameters is specified, the native parameters distribution is first estimated and the new distribution is determined from it:
if the native parameters distribution is normal and the transformation regular at the estimated parameters values: the asymptotic parameters distribution is normal and its covariance matrix determined from the inverse Fisher information matrix of the native parameters and the transformation;
in the other cases, the asymptotic parameters distribution is estimated by Bootstrap on the initial data and kernel fitting.
- getBootstrapSize()¶
Accessor to the bootstrap size.
- Returns:
- sizeint
Size of the bootstrap.
- getClassName()¶
Accessor to the object’s name.
- Returns:
- class_namestr
The object class name (object.__class__.__name__).
- getKnownParameterIndices()¶
Accessor to the known parameters indices.
- Returns:
- indices
Indices Indices of the known parameters.
- indices
- getKnownParameterValues()¶
Accessor to the known parameters values.
- Returns:
- values
Point Values of known parameters.
- values
- getName()¶
Accessor to the object’s name.
- Returns:
- namestr
The name of the object.
- hasName()¶
Test if the object is named.
- Returns:
- hasNamebool
True if the name is not empty.
- setBootstrapSize(bootstrapSize)¶
Accessor to the bootstrap size.
- Parameters:
- sizeint
The size of the bootstrap.
- setKnownParameter(values, positions)¶
Accessor to the known parameters.
- Parameters:
- valuessequence of float
Values of known parameters.
- positionssequence of int
Indices of known parameters.
Examples
When a subset of the parameter vector is known, the other parameters only have to be estimated from data.
In the following example, we consider a sample and want to fit a
Betadistribution. We assume that theand
parameters are known beforehand. In this case, we set the third parameter (at index 2) to -1 and the fourth parameter (at index 3) to 1.
>>> import openturns as ot >>> ot.RandomGenerator.SetSeed(0) >>> distribution = ot.Beta(2.3, 2.2, -1.0, 1.0) >>> sample = distribution.getSample(10) >>> factory = ot.BetaFactory() >>> # set (a,b) out of (r, t, a, b) >>> factory.setKnownParameter([-1.0, 1.0], [2, 3]) >>> inf_distribution = factory.build(sample)
- setName(name)¶
Accessor to the object’s name.
- Parameters:
- namestr
The name of the object.
 OpenTURNS
OpenTURNS