Note
Go to the end to download the full example code.
Create a linear model¶
In this example we create a surrogate model using linear model approximation
with the LinearModelAlgorithm class.
We show how the LinearModelAnalysis class
can be used to produce the statistical analysis of the least squares
regression model.
Introduction¶
The following 2-d function is used in this example:
for any .
Notice that this model is linear:
where ,
and
.
We consider noisy observations of the output:
where with
is the standard deviation.
In our example, we use
.
Finally, we use independent observations of the model.
import openturns as ot
import openturns.viewer as viewer
Simulate the data set¶
We first generate the data and we add noise to the output observations:
ot.RandomGenerator.SetSeed(0)
distribution = ot.Normal(2)
distribution.setDescription(["x", "y"])
sampleSize = 1000
func = ot.SymbolicFunction(["x", "y"], ["3 + 2 * x - y"])
input_sample = distribution.getSample(sampleSize)
epsilon = ot.Normal(0, 0.1).getSample(sampleSize)
output_sample = func(input_sample) + epsilon
Linear regression¶
Let us run the linear model algorithm using the LinearModelAlgorithm
class and get the associated results:
algo = ot.LinearModelAlgorithm(input_sample, output_sample)
result = algo.getResult()
Residuals analysis¶
We can now analyze the residuals of the regression on the training data. For clarity purposes, only the first 5 residual values are printed.
residuals = result.getSampleResiduals()
residuals[:5]
Alternatively, the standardized or studentized residuals can be used:
stdresiduals = result.getStandardizedResiduals()
stdresiduals[:5]
Similarly, we can also obtain the underyling distribution characterizing the residuals:
result.getNoiseDistribution()
Analysis of the results¶
In order to post-process the linear regression results, the LinearModelAnalysis
class can be used:
analysis = ot.LinearModelAnalysis(result)
analysis
The results seem to indicate that the linear model is satisfactory.
The basis uses the three functions
(i.e. the intercept),
and
.
Each row of the table of coefficients tests if one single coefficient is zero. The probability of observing a large value of the T statistics is close to zero for all coefficients. Hence, we can reject the hypothesis that any coefficient is zero. In other words, all the coefficients are significantly nonzero.
The
score is close to 1. Furthermore, the adjusted
value, which takes into account the size of the data set and the number of hyperparameters, is similar to the unadjusted
score. Most of the variance is explained by the linear model.
The F-test tests if all coefficients are simultaneously zero. The Fisher-Snedecor p-value is lower than 1%. Hence, there is at least one nonzero coefficient.
The normality test checks that the residuals have a normal distribution. The normality assumption can be rejected if the p-value is close to zero. The p-values are larger than 0.05: the normality assumption of the residuals is not rejected.
Graphical analyses¶
Let us compare model and fitted values:
graph = analysis.drawModelVsFitted()
view = viewer.View(graph)
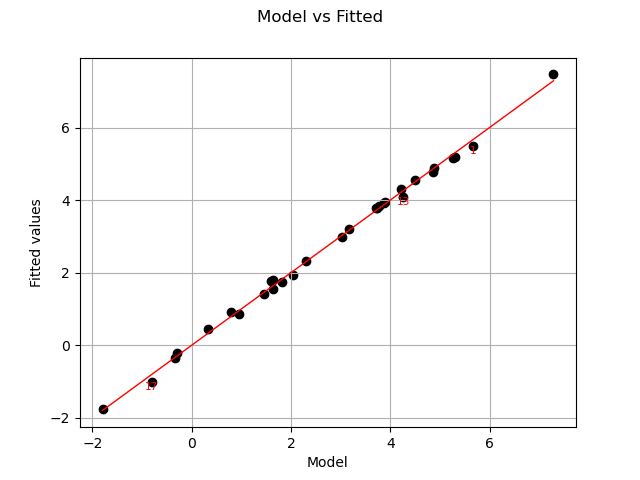
The previous figure seems to indicate that the linearity hypothesis is accurate.
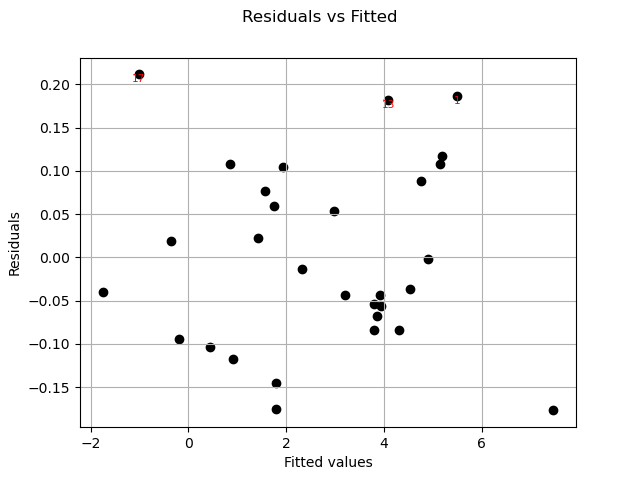
Residuals can be plotted against the fitted values.
graph = analysis.drawResidualsVsFitted()
view = viewer.View(graph)
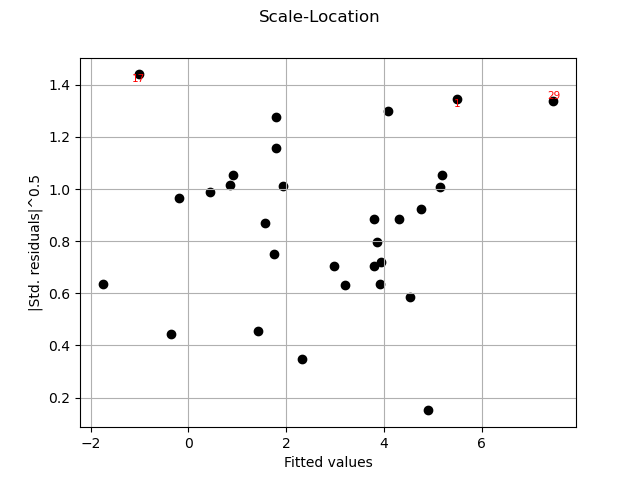
graph = analysis.drawScaleLocation()
view = viewer.View(graph)
graph = analysis.drawQQplot()
view = viewer.View(graph)
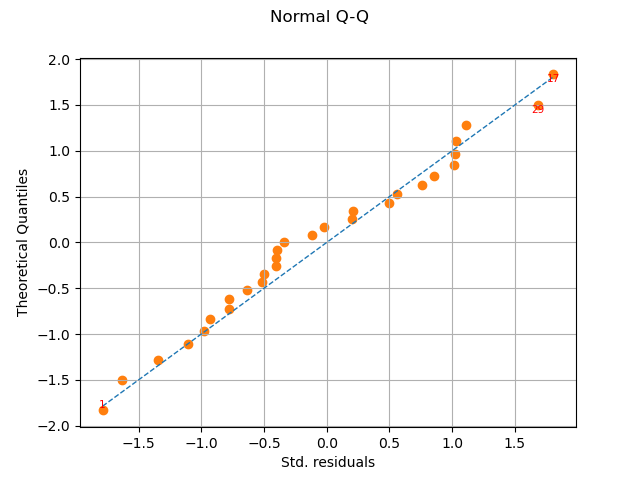
In this case, the two distributions are very close: there is no obvious outlier.
Cook’s distance measures the impact of every individual data point on the linear regression, and can be plotted as follows:
graph = analysis.drawCookDistance()
view = viewer.View(graph)
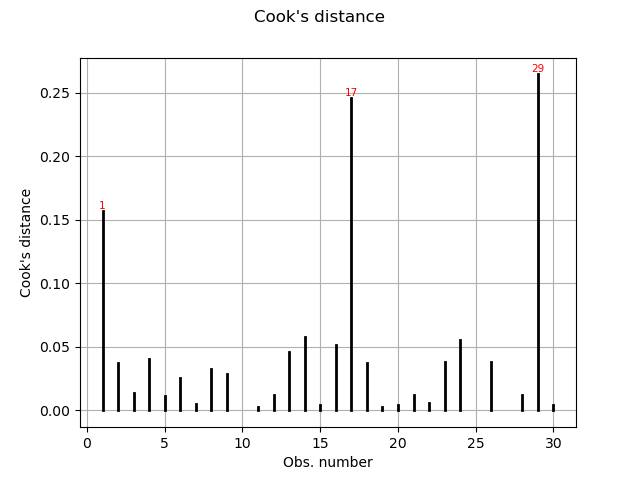
This graph shows us the index of the points with disproportionate influence.
One of the components of the computation of Cook’s distance at a given point is that point’s leverage. High-leverage points are far from their closest neighbors, so the fitted linear regression model must pass close to them.
# sphinx_gallery_thumbnail_number = 6
graph = analysis.drawResidualsVsLeverages()
view = viewer.View(graph)
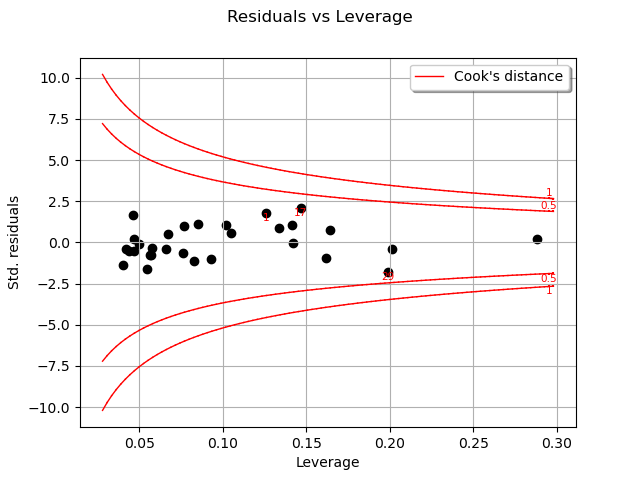
In this case, there seems to be no obvious influential outlier characterized by large leverage and residual values.
Similarly, we can also plot Cook’s distances as a function of the sample leverages:
graph = analysis.drawCookVsLeverages()
view = viewer.View(graph)
![Cook's dist vs Leverage h[ii]/(1-h[ii])](../../_images/sphx_glr_plot_linear_model_007.png)
Finally, we give the intervals for each estimated coefficient (95% confidence interval):
alpha = 0.95
interval = analysis.getCoefficientsConfidenceInterval(alpha)
print("confidence intervals with level=%1.2f: " % (alpha))
print("%s" % (interval))
confidence intervals with level=0.95:
[2.99855, 3.01079]
[1.99374, 2.00616]
[-1.00395, -0.991707]
 OpenTURNS
OpenTURNS