LinearModelValidation¶
(Source code, png)
- class LinearModelValidation(*args)¶
Validate a linear regression metamodel.
Warning
This class is experimental and likely to be modified in future releases. To use it, import the
openturns.experimentalsubmodule.- Parameters:
- result
LinearModelResult A linear model result resulting from linear least squares regression.
- splitter
SplitterImplementation The cross-validation method. The default is
LeaveOneOutSplitter.
- result
Methods
Accessor to the mean squared error.
Compute the R2 score.
Plot a model vs metamodel graph for visual validation.
Accessor to the object's name.
Get the linear model result.
Accessor to the output predictions from the metamodel.
getName()Accessor to the object's name.
Accessor to the output sample.
getResidualDistribution([smooth])Compute the non parametric distribution of the residual sample.
Compute the residual sample.
Get the cross-validation method.
hasName()Test if the object is named.
setName(name)Accessor to the object's name.
See also
Notes
A LinearModelValidation object is used for the validation of a linear model. It is based on the fast (analytical) leave-one-out and fast K-Fold cross-validation methods presented in Validation and cross validation of metamodels.
Analytical cross-validation can only be performed if all coefficients are estimated without model selection: if the coefficients are computed with model selection, then an exception is produced by default. This is because model selection leads to supposedly improved coefficients, so that the hypotheses required to estimate the mean squared error using the cross-validation method are not satisfied anymore. As a consequence, using the analytical formula without taking into account for the model selection leads to a biased, optimistic, mean squared error. More precisely, the analytical formula produces a MSE which is lower than the true one on average. Model selection is involved if the
LinearModelStepwiseAlgorithmclass is involved. If theLinearModelAlgorithmclass is used, then no model selection is involved and the analytical cross-validation methods can be used. If model selection is involved, the naive methods based on theLeaveOneOutSplitterandKFoldSplitterclasses can be used directly, but this can be much slower than the analytical methods implemented in theLinearModelValidationclass. In many cases, however, the order of magnitude of the estimate from the analytical formula applied to a sparse model is correct: the estimate of the MSE is only slightly lower than the true value. In order to enable the calculation of the analytical MSE estimator on a sparse model, please set the LinearModelValidation-ModelSelection key of theResourceMapto False: use this option at your own risks.We suggest to use leave-one-out cross validation when possible, because it produces a more accurate estimate of the error than K-Fold does. If K-Fold is required, we suggest to use the largest possible value of
.
The predictions of the leave-one-one or K-Fold surrogate models are based on the hold-out method. For example, if we use the leave-one-out cross-validation method, the
-th prediction is the prediction of the linear model trained using the hold-out sample where the
-th observation was removed. This produces a sample of residuals which can be retrieved using the
getResidualSamplemethod. ThedrawValidationperforms similarly.Examples
Create a linear model.
>>> import openturns as ot >>> import openturns.experimental as otexp >>> ot.RandomGenerator.SetSeed(0) >>> func = ot.SymbolicFunction( ... ['x1', 'x2', 'x3'], ... ['x1 + x2 + sin(x2 * 2 * pi_) / 5 + 1e-3 * x3^2'] ... ) >>> dimension = 3 >>> distribution = ot.JointDistribution([ot.Normal()] * dimension) >>> sampleSize = 20 >>> inputSample = distribution.getSample(sampleSize) >>> outputSample = func(inputSample) >>> algo = ot.LinearModelAlgorithm(inputSample, outputSample) >>> algo.run() >>> result = algo.getResult()
Validate the linear model using leave-one-out cross-validation.
>>> validation = otexp.LinearModelValidation(result)
We can use a specific cross-validation splitter if needed.
>>> splitterLOO = ot.LeaveOneOutSplitter(sampleSize) >>> validation = otexp.LinearModelValidation(result, splitterLOO) >>> r2Score = validation.computeR2Score() >>> print('R2 = ', r2Score[0]) R2 = 0.98...
Validate the linear model using K-Fold cross-validation.
>>> splitterKFold = ot.KFoldSplitter(sampleSize) >>> validation = otexp.LinearModelValidation(result, splitterKFold)
Validate the linear model using K-Fold cross-validation and set K.
>>> kFoldParameter = 10 >>> splitterKFold = ot.KFoldSplitter(sampleSize, kFoldParameter) >>> validation = otexp.LinearModelValidation(result, splitterKFold)
Draw the validation graph.
>>> graph = validation.drawValidation()
- __init__(*args)¶
- computeMeanSquaredError()¶
Accessor to the mean squared error.
- Returns:
- meanSquaredError
Point The mean squared error of each marginal output dimension.
- meanSquaredError
Notes
The sample mean squared error is:
where
is the sample size,
is the metamodel,
is the input experimental design and
is the output of the model.
If the output is multi-dimensional, the same calculations are repeated separately for each output marginal
for
where
is the output dimension.
- computeR2Score()¶
Compute the R2 score.
- Returns:
- r2Score
Point The coefficient of determination R2
- r2Score
Notes
The coefficient of determination
is the fraction of the variance of the output explained by the metamodel. It is defined as:
where
is the fraction of unexplained variance:
where
is the output of the physical model
,
is the variance of the output and
is the mean squared error of the metamodel:
The sample
is:
where
is the sample size,
is the metamodel,
is the input experimental design,
is the output of the model and
is the sample variance of the output:
where
is the output sample mean:
- drawValidation()¶
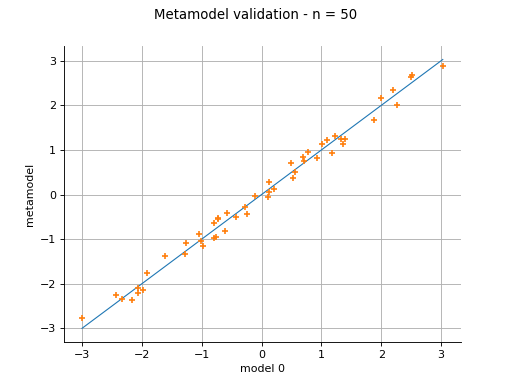
Plot a model vs metamodel graph for visual validation.
- Returns:
- graph
GridLayout The visual validation graph.
- graph
Notes
The plot presents the metamodel predictions depending on the model observations. If the points are close to the diagonal line of the plot, then the metamodel validation is satisfactory. Points which are far away from the diagonal represent outputs for which the metamodel is not accurate.
If the output is multi-dimensional, the graph has 1 row and
columns, where
is the output dimension.
- getClassName()¶
Accessor to the object’s name.
- Returns:
- class_namestr
The object class name (object.__class__.__name__).
- getLinearModelResult()¶
Get the linear model result.
- Returns:
- linearModelResult
LinearModelResult The linear model result.
- linearModelResult
- getMetamodelPredictions()¶
Accessor to the output predictions from the metamodel.
- Returns:
- outputMetamodelSample
Sample Output sample of the metamodel.
- outputMetamodelSample
- getName()¶
Accessor to the object’s name.
- Returns:
- namestr
The name of the object.
- getOutputSample()¶
Accessor to the output sample.
- Returns:
- outputSample
Sample Output sample of a model evaluated apart.
- outputSample
- getResidualDistribution(smooth=True)¶
Compute the non parametric distribution of the residual sample.
- Parameters:
- smoothbool
Tells if distribution is smooth (true) or not. Default argument is true.
- Returns:
- residualDistribution
Distribution The residual distribution.
- residualDistribution
Notes
The residual distribution is built thanks to
KernelSmoothingif smooth argument is true. Otherwise, an histogram distribution is returned, thanks toHistogramFactory.
- getResidualSample()¶
Compute the residual sample.
- Returns:
- residual
Sample The residual sample.
- residual
Notes
The residual sample is given by :
for
where
is the sample size,
is the model observation,
is the metamodel and
is the
-th input observation.
If the output is multi-dimensional, the residual sample has dimension
, where
is the output dimension.
- getSplitter()¶
Get the cross-validation method.
- Returns:
- splitter
SplitterImplementation The cross-validation method.
- splitter
- hasName()¶
Test if the object is named.
- Returns:
- hasNamebool
True if the name is not empty.
- setName(name)¶
Accessor to the object’s name.
- Parameters:
- namestr
The name of the object.
 OpenTURNS
OpenTURNS