Note
Go to the end to download the full example code.
FMUPointToFieldFunction basics
otfmi.FMUPointToFieldFunction wraps the FMU into an
openturns.PointToFieldFunction object.
This kind of function accepts openturns.Point or
openturns.Sample as inputs, and outputs a
openturns.Sample or a set of openturns.Field.
First, retrieve the path to epid.fmu. Recall the deviation model is dynamic, i.e. its output evolves over time.
import openturns as ot
import otfmi.example.utility
import matplotlib.pyplot as plt
import openturns.viewer as viewer
path_fmu = otfmi.example.utility.get_path_fmu("epid")
Define the time grid for the FMU’s output. The last value of the time grid, here 10., will define the FMU stop time for simulation.
mesh = ot.RegularGrid(0.0, 0.1, 2000)
meshSample = mesh.getVertices()
print(meshSample)
[ t ]
0 : [ 0 ]
1 : [ 0.1 ]
2 : [ 0.2 ]
...
1997 : [ 199.7 ]
1998 : [ 199.8 ]
1999 : [ 199.9 ]
Note
The FMU solver uses its own time grid for simulation. The FMU output is then interpolated on the user-provided time grid.
Wrap the FMU in an openturns.PointToFieldFunction object:
function = otfmi.FMUPointToFieldFunction(
mesh,
path_fmu,
inputs_fmu=["infection_rate"],
outputs_fmu=["infected"],
start_time=0.0,
final_time=200.0,
)
print(type(function))
<class 'openturns.func.PointToFieldFunction'>
Note
The start and final times must define an interval comprising the mesh. Setting manually the start and final times is recommended to avoid uncontrolled simulation duration.
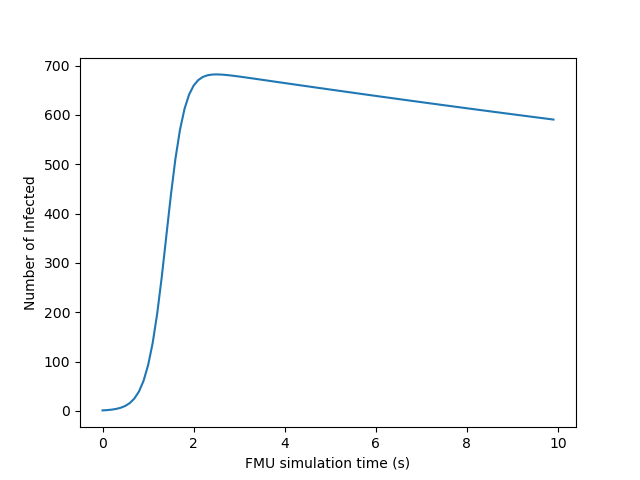
Simulate the function on an input openturns.Point yields an output
openturns.Sample, corresponding to the output evolution over time:
inputPoint = ot.Point([2.0])
outputSample = function(inputPoint)
plt.xlabel("FMU simulation time (s)")
plt.ylabel("Number of Infected")
plt.plot(meshSample, outputSample)
plt.show()
Simulate the function on a input openturns.Sample yields a set of
fields called openturns.ProcessSample:
inputSample = ot.Sample([[2.0], [2.25], [2.5]])
outputProcessSample = function(inputSample)
print(outputProcessSample)
[field 0:
[ t infected ]
0 : [ 0 1 ]
1 : [ 0.1 1.14974 ]
2 : [ 0.2 1.29948 ]
...
1997 : [ 199.7 1.46173e-42 ]
1998 : [ 199.8 1.38833e-42 ]
1999 : [ 199.9 1.31492e-42 ]
field 1:
[ t infected ]
0 : [ 0 1 ]
1 : [ 0.1 1.17471 ]
2 : [ 0.2 1.34941 ]
...
1997 : [ 199.7 7.73327e-44 ]
1998 : [ 199.8 7.33573e-44 ]
1999 : [ 199.9 6.9382e-44 ]
field 2:
[ t infected ]
0 : [ 0 1 ]
1 : [ 0.1 1.19967 ]
2 : [ 0.2 1.39934 ]
...
1997 : [ 199.7 1.38086e-44 ]
1998 : [ 199.8 1.30896e-44 ]
1999 : [ 199.9 1.23707e-44 ]]
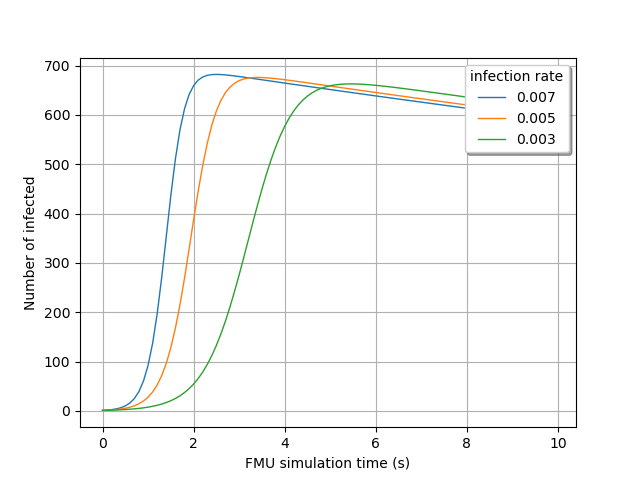
Visualize the time evolution of the infected over time, depending on the
ìnfection_rate` value:
gridLayout = outputProcessSample.draw()
graph = gridLayout.getGraph(0, 0)
graph.setTitle("")
graph.setXTitle("FMU simulation time (s)")
graph.setYTitle("Number of infected")
graph.setLegends([str(line[0]) for line in inputSample])
view = viewer.View(graph, legend_kw={"title": "infection rate"})
view.ShowAll()
Total running time of the script: (0 minutes 0.247 seconds)