Note
Go to the end to download the full example code.
Example 1: Axial stressed beam¶
In this test case, we create a sample from a mixture and we try to estimate the mixture parameters from the sample. It is not a really an example of a study but it shows how to use this module. The optimal number of clusters is not supposed to be known, and will be estimated as well. We are in dimension 2, and the reference mixture is defined from 3 normal distributions:
with:
with
,
,
and
,
with
,
,
and
,
with
,
,
and
.
import openturns as ot
import openturns.viewer as otv
import otmixmod
Create a multidimensional sample from a mixture of Normal
dim = 2
size = 20000
coll = []
R = ot.CorrelationMatrix(dim)
First atom
Second atom
Third atom
Reference mixture
mixture = ot.Mixture(coll, weights)
Creation of the numerical Sample from which we will estimate the parameters of the mixture.
sample = mixture.getSample(size)
Creation of the mixture factory
myAtomsNumber = 3
myCovModel = 'Gaussian_pk_L_Dk_A_Dk'
bestLL = -1e100
bestMixture = ot.Mixture()
bestNbClusters = 0
stop = False
nbClusters = 1
while not stop:
factory = otmixmod.MixtureFactory(nbClusters, myCovModel)
# Estimation of the parameters of the mixture
estimatedDistribution, labels, logLikelihood = factory.build(sample)
stop = logLikelihood[1] <= bestLL
if not stop:
bestLL = logLikelihood[1]
bestNbClusters = nbClusters
bestMixture = estimatedDistribution
nbClusters += 1
print("best number of atoms=", bestNbClusters)
myAtomsNumber = bestNbClusters
estimatedDistribution = bestMixture
# Some printings to show the result
print("Covariance Model used=", myCovModel)
print("")
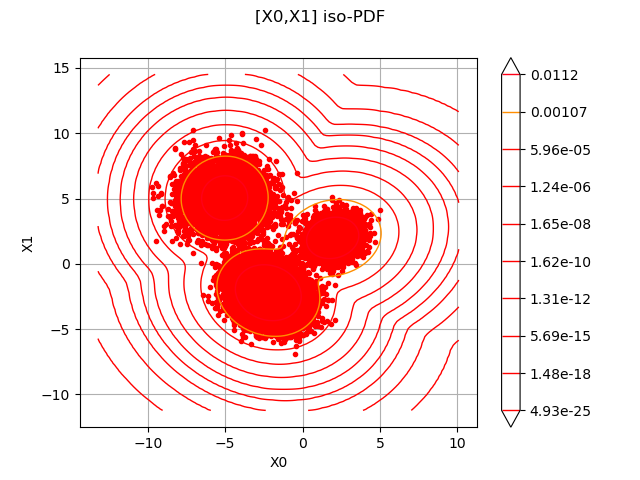
print("Estimated distribution:", estimatedDistribution)
best number of atoms= 3
Covariance Model used= Gaussian_pk_L_Dk_A_Dk
Estimated distribution: Mixture((w = 0.254333, d = Normal(mu = [1.94368,1.98439], sigma = [1.21056,1.13791], R = [[ 1 0.119447 ]
[ 0.119447 1 ]])), (w = 0.50104, d = Normal(mu = [-2.21622,-2.21449], sigma = [1.17175,1.17784], R = [[ 1 -0.134189 ]
[ -0.134189 1 ]])), (w = 0.244627, d = Normal(mu = [-5.03441,5.03529], sigma = [1.09358,1.25075], R = [[ 1 0.0148212 ]
[ 0.0148212 1 ]])))
Some drawings
if sample.getDimension() == 2:
g = estimatedDistribution.drawPDF()
c = ot.Cloud(sample)
c.setColor("red")
c.setPointStyle("bullet")
ctmp = g.getDrawable(0)
g.setDrawable(c, 0)
g.add(ctmp)
view = otv.View(g)
otv.View.ShowAll()
Total running time of the script: (0 minutes 1.364 seconds)